Variations in Soil Enzyme Activities and Microbial Communities along an Altitudinal Gradient on the Eastern Qinghai–Tibetan Plateau
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Sampling
2.2. Soil Chemical Properties
2.3. Soil Enzyme Activity
2.4. DNA Extraction, PCR and High-Throughput Sequencing
2.5. Data Analysis
3. Results
3.1. Characteristics of Soil Chemical Properties
3.2. Spatial Variations in Soil Enzyme Activities
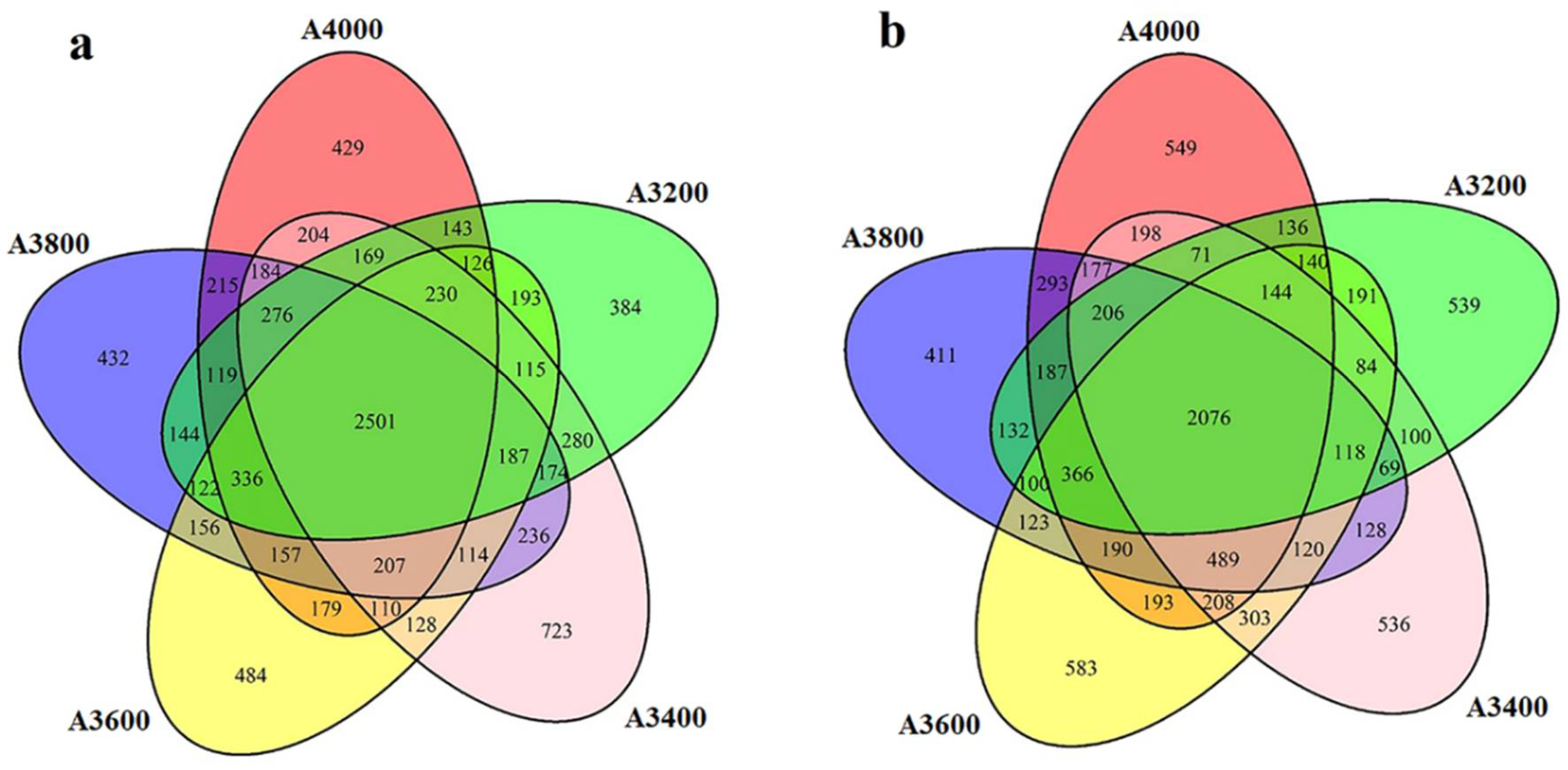
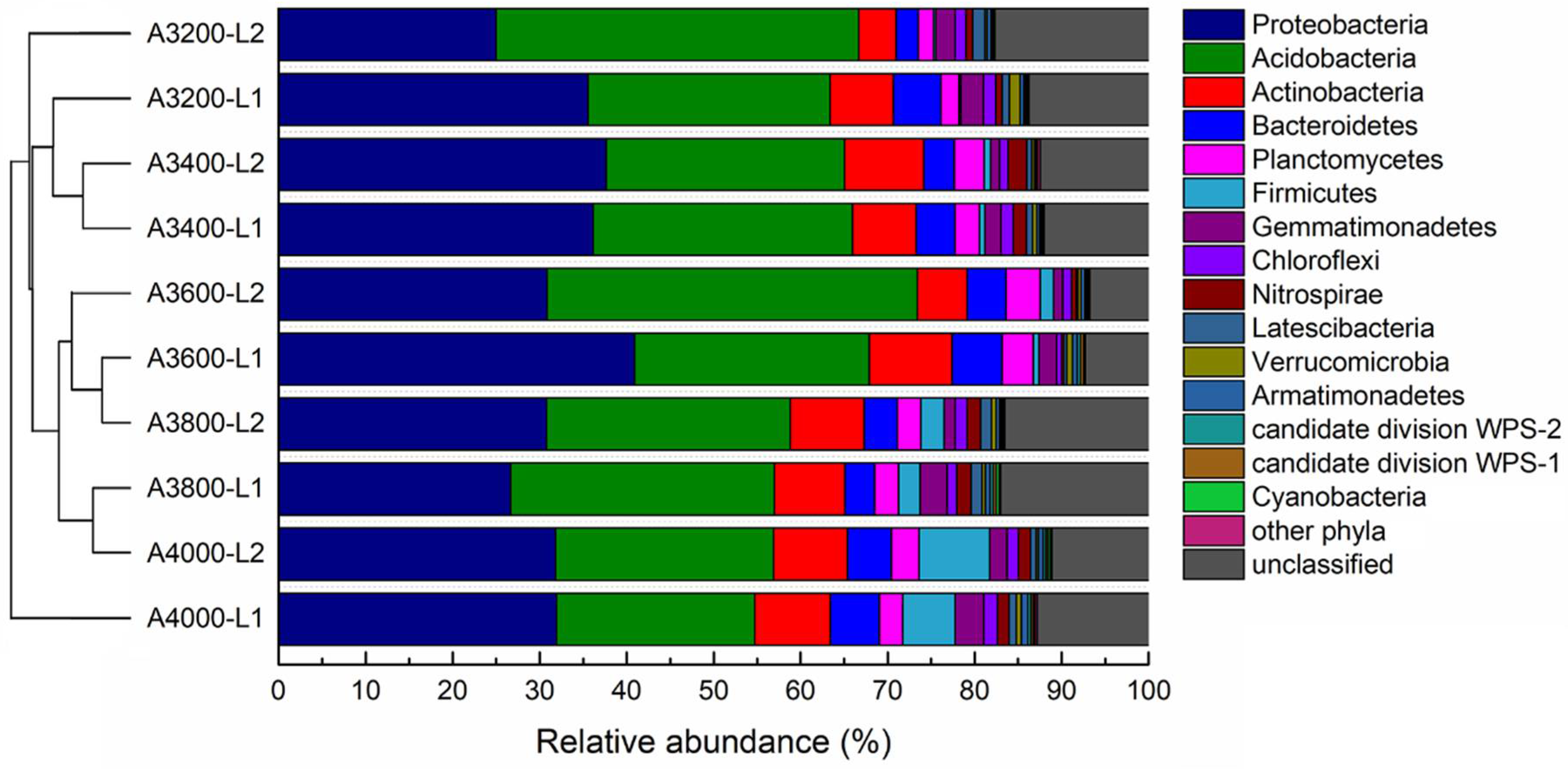
3.3. Composition of Soil Microbial Communities
3.4. Diversity Indices of Soil Microbial Communities
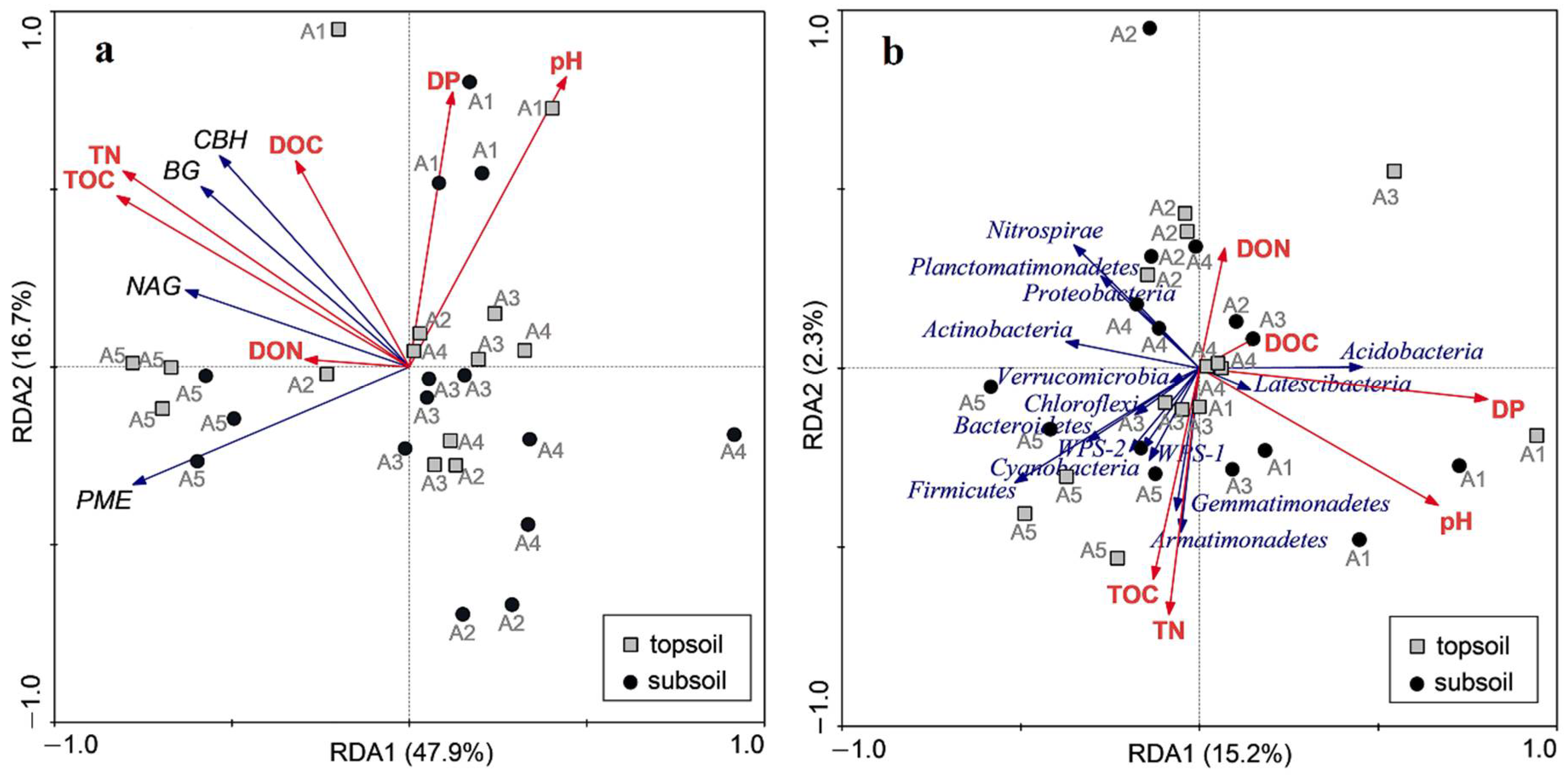
3.5. Correlations between Soil Chemical Properties, Enzyme Activities and Microbial Communities
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, H.; Zhu, Q.; Peng, C.; Wu, N.; Wang, Y.; Fang, X.; Gao, Y.; Zhu, D.; Yang, G.; Tian, J.; et al. The impacts of climate change and human activities on biogeochemical cycles on the Qinghai-Tibetan Plateau. Glob. Chang. Biol. 2013, 19, 2940–2955. [Google Scholar] [CrossRef]
- Wang, G.X.; Qian, J.; Cheng, G.D.; Lai, Y.M. Soil organic carbon pool of grassland soils on the Qinghai-Tibetan Plateau and its global implication. Sci. Total Environ. 2002, 291, 207–217. [Google Scholar] [CrossRef]
- Gao, Q.; Guo, Y.; Xu, H.; Ganjurjav, H.; Li, Y.; Wan, Y.; Qin, X.; Ma, X.; Liu, S. Climate change and its impacts on vegetation distribution and net primary productivity of the alpine ecosystem in the Qinghai-Tibetan Plateau. Sci. Total Environ. 2016, 554, 34–41. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Gao, Q.; Liu, S.; Zhou, H.; Ganjurjav, H.; Wang, X. Climate change and human activities altered the diversity and composition of soil microbial community in alpine grasslands of the Qinghai-Tibetan Plateau. Sci. Total Environ. 2016, 562, 353–363. [Google Scholar] [CrossRef]
- Mu, C.; Zhang, T.; Zhang, X.; Cao, B.; Peng, X.; Cao, L.; Su, H. Pedogenesis and physicochemical parameters influencing soil carbon and nitrogen of alpine meadows in permafrost regions in the northeastern Qinghai-Tibetan Plateau. Catena 2016, 141, 85–91. [Google Scholar] [CrossRef]
- Mu, C.C.; Abbott, B.W.; Zhao, Q.; Su, H.; Wang, S.F.; Wu, Q.B.; Zhang, T.J.; Wu, X.D. Permafrost collapse shifts alpine tundra to a carbon source but reduces N2O and CH4 release on the northern Qinghai-Tibetan Plateau. Geophys. Res. Lett. 2017, 44, 8945–8952. [Google Scholar] [CrossRef]
- Wu, X.; Fang, H.; Zhao, Y.; Smoak, J.M.; Li, W.; Shi, W.; Sheng, Y.; Zhao, L.; Ding, Y. A conceptual model of the controlling factors of soil organic carbon and nitrogen densities in a permafrost-affected region on the eastern Qinghai-Tibetan Plateau. J. Geophys. Res. Biogeosciences 2017, 122, 1705–1717. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Zhang, X.; Shi, S.; Cao, W. Effect of degradation and rebuilding of artificial grasslands on soil respiration and carbon and nitrogen pools on an alpine meadow of the Qinghai-Tibetan Plateau. Ecol. Eng. 2018, 111, 134–142. [Google Scholar] [CrossRef]
- Ma, L.; Yao, Z.; Zheng, X.; Zhang, H.; Wang, K.; Zhu, B.; Wang, R.; Zhang, W.; Liu, C. Increasing grassland degradation stimulates the non-growing season CO2 emissions from an alpine meadow on the Qinghai-Tibetan Plateau. Environ. Sci. Pollut. Res. 2018, 25, 26576–26591. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, H.; Cui, L.; Wickings, K.; Fu, S.; Wang, C. Impacts of alpine wetland degradation on the composition, diversity and trophic structure of soil nematodes on the Qinghai-Tibetan Plateau. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.Y.; Yan, Y.; Sun, J.; Zhang, X.K.; Chen, Y.C.; Wang, X.D.; Cheng, G.W. Carbon, nitrogen, and phosphorus storage in alpine grassland ecosystems of Tibet: Effects of grazing exclusion. Ecol. Evol. 2015, 5, 4492–4504. [Google Scholar] [CrossRef]
- Xu, W.; Ma, L.; Ma, M.; Zhang, H.; Yuan, W. Spatial-temporal variability of snow cover and depth in the Qinghai-Tibetan Plateau. J. Clim. 2017, 30, 1521–1533. [Google Scholar] [CrossRef]
- He, D.; Xiang, X.; He, J.-S.; Wang, C.; Cao, G.; Adams, J.; Chu, H. Composition of the soil fungal community is more sensitive to phosphorus than nitrogen addition in the alpine meadow on the Qinghai-Tibetan Plateau. Biol. Fertil. Soils 2016, 52, 1059–1072. [Google Scholar] [CrossRef]
- Siles, J.A.; Cajthaml, T.; Minerbi, S.; Margesin, R. Effect of altitude and season on microbial activity, abundance and community structure in Alpine forest soils. FEMS Microbiol. Ecol. 2016, 92, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.X.; He, X.H.; Wu, F.Z.; Yang, W.Q.; Tan, B. Decomposition of Abies faxoniana litter varies with freeze-thaw stages and altitudes in subalpine/alpine forests of southwest China. Scand. J. For. Res. 2012, 27, 586–596. [Google Scholar] [CrossRef]
- Bruun, H.H.; Moen, J.; Virtanen, R.; Grytnes, J.A.; Oksanen, L.; Angerbjorn, A. Effects of altitude and topography on species richness of vascular plants, bryophytes and lichens in alpine communities. J. Veg. Sci. 2006, 17, 37–46. [Google Scholar] [CrossRef]
- Withington, C.L.; Sanford, R.L., Jr. Decomposition rates of buried substrates increase with altitude in the forest-alpine tundra ecotone. Soil Biol. Biochem. 2007, 39, 68–75. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, D.; Sun, D.; Wang, X.; Smith, J.L.; Du, G. Impacts of altitude and position on the rates of soil nitrogen mineralization and nitrification in alpine meadows on the eastern Qinghai-Tibetan Plateau, China. Biol. Fertil. Soils 2012, 48, 393–400. [Google Scholar] [CrossRef]
- Jha, D.K.; Sharma, G.D.; Mishra, R.R. Soil microbial population numbers and enzyme activities in relation to altitude and forest degradation. Soil Biol. Biochem. 1992, 24, 761–767. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, G.; Zhang, X.; He, N.; Wang, Q.; Wang, S.; Wang, R.; Zhao, N.; Jia, Y.; Wang, C. Soil enzyme activity and stoichiometry in forest ecosystems along the North-South Transect in eastern China (NSTEC). Soil Biol. Biochem. 2017, 104, 152–163. [Google Scholar] [CrossRef]
- Trasar-Cepeda, C.; Leiros, M.C.; Seoane, S.; Gil-Sotres, F. Limitations of soil enzymes as indicators of soil pollution. Soil Biol. Biochem. 2000, 32, 1867–1875. [Google Scholar] [CrossRef]
- Deng, S.P.; Tabatabai, M.A. Cellulase activity of soils. Soil Biol. Biochem. 1994, 26, 1347–1354. [Google Scholar] [CrossRef]
- Jian, S.; Li, J.; Chen, J.; Wang, G.; Mayes, M.A.; Dzantor, K.E.; Hui, D.; Luo, Y. Soil extracellular enzyme activities, soil carbon and nitrogen storage under nitrogen fertilization: A meta-analysis. Soil Biol. Biochem. 2016, 101, 32–43. [Google Scholar] [CrossRef] [Green Version]
- Hui, D.; Mayes, M.A.; Wang, G. Kinetic parameters of phosphatase: A quantitative synthesis. Soil Biol. Biochem. 2013, 65, 105–113. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Ren, G.; Teng, Y.; Ren, W.; Dai, S.; Li, Z. Pyrene dissipation potential varies with soil type and associated bacterial community changes. Soil Biol. Biochem. 2016, 103, 71–85. [Google Scholar] [CrossRef]
- Ren, G.; Ren, W.; Teng, Y.; Li, Z. Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Nielsen, S.; Minchin, T.; Kimber, S.; van Zwieten, L.; Gilbert, J.; Munroe, P.; Joseph, S.; Thomas, T. Comparative analysis of the microbial communities in agricultural soil amended with enhanced biochars or traditional fertilisers. Agric. Ecosyst. Environ. 2014, 191, 73–82. [Google Scholar] [CrossRef]
- Guo, W.; Andersen, M.N.; Qi, X.-B.; Li, P.; Li, Z.-Y.; Fan, X.-Y.; Zhou, Y. Effects of reclaimed water irrigation and nitrogen fertilization on the chemical properties and microbial community of soil. J. Integr. Agric. 2017, 16, 679–690. [Google Scholar] [CrossRef]
- Lu, L.; Han, W.; Zhang, J.; Wu, Y.; Wang, B.; Lin, X.; Zhu, J.; Cai, Z.; Jia, Z. Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J. 2012, 6, 1978–1984. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Jiang, Y.; Wang, F.; Wen, C.; Deng, Y.; Xue, K.; Qin, Y.; Yang, Y.; Wu, L.; Zhou, J.; et al. Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. ISME J. 2015, 9, 2561–2572. [Google Scholar] [CrossRef] [Green Version]
- Meola, M.; Lazzaro, A.; Zeyer, J. Diversity, resistance and resilience of the bacterial communities at two alpine glacier forefields after a reciprocal soil transplantation. Environ. Microbiol. 2014, 16, 1918–1934. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Hu, W.; Tao, J.; Liu, Y.; Kong, Z.; Wu, L. Soil bacterial community structure and extracellular enzyme activities under different land use types in a long-term reclaimed wetland. J. Soils Sediments 2019, 19, 2543–2557. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Shah, J.J.F. Ecoenzymatic stoichiometry of recalcitrant organic matter decomposition: The growth rate hypothesis in reverse. Biogeochemistry 2011, 102, 31–43. [Google Scholar] [CrossRef]
- Steinweg, J.M.; Dukes, J.S.; Paul, E.A.; Wallenstein, M.D. Microbial responses to multi-factor climate change: Effects on soil enzymes. Front. Microbiol. 2013, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Jones, D.L.; Willett, V.B. Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biol. Biochem. 2006, 38, 991–999. [Google Scholar] [CrossRef]
- Dick, R.P. Methods of Soil Enzymology; Soil Science Society of America: Madison, WI, USA, 2011. [Google Scholar]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Bacci, G.; Bani, A.; Bazzicalupo, M.; Ceccherini, M.T.; Galardini, M.; Nannipieri, P.; Pietramellara, G.; Mengoni, A. Evaluation of the performances of ribosomal database project (RDP) classifier for taxonomic assignment of 16S rRNA metabarcoding sequences generated from Illumina-Solexa NGS. J Genom. 2015, 3, 36–39. [Google Scholar] [CrossRef] [Green Version]
- Gaston, K.J. Global patterns in biodiversity. Nature 2000, 405, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Q.; Ouyang, H.; Lei, G.C.; Xu, X.L.; Zhang, M.X. Effects of temperature, soil moisture, soil type and their interactions on soil carbon mineralization in Zoig alpine wetland, Qinghai-Tibet Plateau. Chin. Geogr. Sci. 2011, 21, 27–35. [Google Scholar] [CrossRef]
- Weedon, J.T.; Aerts, R.; Kowalchuk, G.A.; van Logtestijn, R.; Andringa, D.; van Bodegom, P.M. Temperature sensitivity of peatland C and N cycling: Does substrate supply play a role? Soil Biol. Biochem. 2013, 61, 109–120. [Google Scholar] [CrossRef]
- Waring, B.G.; Weintraub, S.R.; Sinsabaugh, R.L. Ecoenzymatic stoichiometry of microbial nutrient acquisition in tropical soils. Biogeochemistry 2014, 117, 101–113. [Google Scholar] [CrossRef]
- Olander, L.P.; Vitousek, P.M. Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry 2000, 49, 175–190. [Google Scholar] [CrossRef]
- Ren, C.; Zhao, F.; Shi, Z.; Chen, J.; Han, X.; Yang, G.; Feng, Y.; Ren, G. Differential responses of soil microbial biomass and carbon-degrading enzyme activities to altered precipitation. Soil Biol. Biochem. 2017, 115, 1–10. [Google Scholar] [CrossRef]
- Chu, H.; Fierer, N.; Lauber, C.L.; Caporaso, J.G.; Knight, R.; Grogan, P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 2010, 12, 2998–3006. [Google Scholar] [CrossRef]
- Kui, L.; Sun, H.; Lei, Q.; Gao, W.; Bao, L.; Chen, Y.; Jia, Z. Soil microbial community assemblage and its seasonal variability in alpine treeline ecotone on the eastern Qinghai-Tibet Plateau. Soil Ecol. Lett. 2019, 1, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Ma, A.; Qi, H.; Zhuang, X.; Zhuang, G.; Zhao, G. Warmer temperature accelerates methane emissions from the Zoige wetland on the Tibetan Plateau without changing methanogenic community composition. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Sinsabaugh, R.L.; Lauber, C.L.; Weintraub, M.N.; Ahmed, B.; Allison, S.D.; Crenshaw, C.; Contosta, A.R.; Cusack, D.; Frey, S.; Gallo, M.E.; et al. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Liptzin, D. C:N:P stoichiometry in soil: Is there a “Redfield ratio” for the microbial biomass? Biogeochemistry 2007, 85, 235–252. [Google Scholar] [CrossRef]
- Rumpel, C.; Kogel-Knabner, I. Deep soil organic matter-a key but poorly understood component of terrestrial C cycle. Plant Soil 2011, 338, 143–158. [Google Scholar] [CrossRef]
- Fontaine, S.; Barot, S.; Barre, P.; Bdioui, N.; Mary, B.; Rumpel, C. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 2007, 450, 277–280. [Google Scholar] [CrossRef] [PubMed]
- de Graaff, M.A.; Jastrow, J.D.; Gillette, S.; Johns, A.; Wullschleger, S.D. Differential priming of soil carbon driven by soil depth and root impacts on carbon availability. Soil Biol. Biochem. 2014, 69, 147–156. [Google Scholar] [CrossRef]
- Niemi, R.M.; Vepsalainen, M.; Wallenius, K.; Simpanen, S.; Alakukku, L.; Pietola, L. Temporal and soil depth-related variation in soil enzyme activities and in root growth of red clover (Trifolium pratense) and timothy (Phleum pratense) in the field. Appl. Soil Ecol. 2005, 30, 113–125. [Google Scholar] [CrossRef]
- Agnelli, A.; Ascher, J.; Corti, G.; Ceccherini, M.T.; Nannipieri, P.; Pietramellara, G. Distribution of microbial communities in a forest soil profile investigated by microbial biomass, soil respiration and DGGE of total and extracellular DNA. Soil Biol. Biochem. 2004, 36, 859–868. [Google Scholar] [CrossRef]
- Jia, J.; Feng, X.; He, J.-S.; He, H.; Lin, L.; Liu, Z. Comparing microbial carbon sequestration and priming in the subsoil versus topsoil of a Qinghai-Tibetan alpine grassland. Soil Biol. Biochem. 2017, 104, 141–151. [Google Scholar] [CrossRef]


| Altitude ID | Community Type | Sampling Altitude (m) | Longitude (E) | Latitude (N) |
|---|---|---|---|---|
| A3200 | Alpine meadow | 3237 | 102°25′ | 31°32′ |
| A3400 | Treeline Ecotone | 3430 | 102°23′ | 31°36′ |
| A3600 | Alpine forest | 3606 | 102°20′ | 31°39′ |
| A3800 | Alpine forest | 3810 | 102°19′ | 31°41′ |
| A4000 | Alpine forest | 4018 | 102°19′ | 31°42′ |
| Soil Layer | Altitude ID | pH | TOC | TN | DOC | DON | DP |
|---|---|---|---|---|---|---|---|
| (in Water) | (g/kg) | (g/kg) | (g/kg) | (g/kg) | (mg/kg) | ||
| Topsoil (0–10 cm) | A3200 | 7.78 ± 0.02a | 65.57 ± 9.42b | 6.82 ± 1.04b | 0.81 ± 0.16 | 0.14 ± 0.02 | 13.33 ± 1.89a |
| A3400 | 5.89 ± 0.48c | 65.29 ± 8.66b | 5.64 ± 0.82bc | 0.74 ± 0.10 | 0.21 ± 0.04 | 10.10 ± 0.95b | |
| A3600 | 6.72 ± 0.39b | 51.86 ± 4.98c | 5.55 ± 0.65bc | 0.56 ± 0.10 | 0.24 ± 0.20 | 8.33 ± 1.62bc | |
| A3800 | 6.66 ± 0.45b | 40.38 ± 4.27c | 4.92 ± 0.45c | 0.60 ± 0.06 | 0.14 ± 0.02 | 8.27 ± 0.60bc | |
| A4000 | 5.92 ± 0.11c | 85.24 ± 3.03a | 8.21 ± 0.34a | 0.64 ± 0.06 | 0.20 ± 0.02 | 7.63 ± 0.64c | |
| Mean | 6.60 ± 0.20 | 61.67 ± 4.27 | 6.23 ± 0.35 | 0.67 ± 0.03 | 0.18 ± 0.02 | 9.53 ± 0.62 | |
| Subsoil (10–20 cm) | A3200 | 7.80 ± 0.03A | 68.33 ± 5.59A | 6.80 ± 0.44AB | 0.67 ± 0.10A | 0.14 ± 0.02AB | 12.03 ± 2.34A |
| A3400 | 5.40 ± 0.71C | 35.47 ± 14.54C | 3.71 ± 1.40C | 0.62 ± 0.11AB | 0.17 ± 0.04AB | 8.50 ± 1.10B | |
| A3600 | 6.64 ± 0.21B | 49.83 ± 2.13B | 5.49 ± 0.15B | 0.51 ± 0.04BC | 0.15 ± 0.07AB | 7.80 ± 0.89B | |
| A3800 | 6.80 ± 0.55B | 25.69 ± 7.85C | 3.39 ± 0.83C | 0.47 ± 0.06C | 0.10 ± 0.03B | 6.23 ± 0.21B | |
| A4000 | 5.76 ± 0.22C | 77.25 ± 0.44A | 7.38 ± 0.18A | 0.65 ± 0.02AB | 0.21 ± 0.01A | 7.50 ± 1.42B | |
| Mean | 6.48 ± 0.23 | 51.35 ± 5.45 | 5.35 ± 0.46 | 0.58 ± 0.03 | 0.15 ± 0.01 | 8.41 ± 0.60 |
| Soil Layer | Altitude ID | Total Reads | OTU Taxa | Richness | Evenness | Diversity | |
|---|---|---|---|---|---|---|---|
| Chao1 | Pielou | Shannon | Simpson | ||||
| Topsoil (0–10 cm) | A3200 | 48,612 | 3946 | 5687 | 0.789 | 6.529 | 0.993 |
| A3400 | 37,415 | 3393 | 4979 | 0.781 | 6.342 | 0.992 | |
| A3600 | 37,965 | 3476 | 5083 | 0.814 | 6.632 | 0.995 | |
| A3800 | 37,291 | 3198 | 4741 | 0.786 | 6.340 | 0.992 | |
| A4000 | 37,684 | 3473 | 5055 | 0.811 | 6.609 | 0.995 | |
| Mean ± SE | 39,163 ± 2298 | 3465 ± 113 | 5068 ± 141 | 0.797 ± 0.008 | 6.487 ± 0.075 | 0.994 ± 0.001 | |
| Subsoil (10–20 cm) | A3200 | 29,441 | 2577 | 3983 | 0.763 | 5.930 | 0.987 |
| A3400 | 35,860 | 3360 | 4976 | 0.797 | 6.467 | 0.994 | |
| A3600 | 41,197 | 2904 | 4522 | 0.744 | 5.931 | 0.989 | |
| A3800 | 34,259 | 2998 | 4514 | 0.793 | 6.338 | 0.993 | |
| A4000 | 38,054 | 3341 | 4868 | 0.791 | 6.420 | 0.993 | |
| Mean ± SE | 35,762 ± 2802 | 3036 ± 170 | 4572 ± 211 | 0.778 ± 0.001 | 6.217 ± 0.107 | 0.991 ± 0.001 | |
| Soil Layer | Parameter | pH | TOC | TN | DOC | DON | DP | BG | CBH | NAG | PME | Total Reads | OTU Taxa | Chao1 | Pielou | Shannon |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Topsoil (10–20 cm) | TOC | −0.295 | ||||||||||||||
| TN | −0.103 | 0.933 ** | ||||||||||||||
| DOC | 0.251 | 0.472 | 0.441 | |||||||||||||
| DON | −0.187 | 0.251 | 0.242 | 0.231 | ||||||||||||
| DP | 0.489 | 0.047 | −0.014 | 0.636 * | 0.123 | |||||||||||
| BG | −0.285 | 0.585 * | 0.670 ** | 0.087 | −0.101 | −0.376 | ||||||||||
| CBH | 0.054 | 0.440 | 0.638 * | 0.228 | −0.147 | −0.174 | 0.734 ** | |||||||||
| NAG | −0.530 | 0.484 | 0.426 | 0.209 | 0.032 | −0.137 | 0.626 * | 0.347 | ||||||||
| PME | −0.691 ** | 0.583 * | 0.551 * | −0.130 | 0.030 | −0.483 | 0.766 ** | 0.465 | 0.771 ** | |||||||
| Total Reads | 0.282 | 0.028 | 0.041 | 0.212 | 0.218 | 0.670 ** | −0.179 | 0.062 | −0.227 | −0.337 | ||||||
| OTU Taxa | 0.357 | 0.126 | 0.104 | 0.069 | −0.005 | 0.415 | −0.170 | 0.030 | −0.517 | −0.412 | 0.719 ** | |||||
| Chao1 | 0.394 | 0.106 | 0.085 | 0.058 | −0.050 | 0.420 | −0.197 | 0.018 | −0.532 | −0.433 | 0.699 ** | 0.995 ** | ||||
| Pielou | 0.125 | 0.061 | 0.121 | −0.397 | −0.192 | −0.529 | 0.119 | 0.039 | −0.414 | 0.005 | −0.458 | 0.221 | 0.237 | |||
| Shannon | 0.218 | 0.105 | 0.150 | −0.317 | −0.159 | −0.328 | 0.052 | 0.051 | −0.529 | −0.130 | −0.157 | 0.530 | 0.541 * | 0.944 ** | ||
| Simpson | 0.148 | 0.123 | 0.179 | −0.370 | −0.081 | −0.441 | 0.102 | 0.080 | −0.465 | −0.017 | −0.299 | 0.361 | 0.373 | 0.975 ** | 0.970 ** | |
| Subsoil (10–20 cm) | TOC | 0.185 | ||||||||||||||
| TN | 0.264 | 0.990 ** | ||||||||||||||
| DOC | −0.058 | 0.527 * | 0.460 | |||||||||||||
| DON | −0.484 | 0.461 | 0.400 | 0.560 * | ||||||||||||
| DP | 0.503 | 0.449 | 0.440 | 0.661 ** | 0.106 | |||||||||||
| BG | 0.316 | 0.831 ** | 0.816 ** | 0.588 * | 0.325 | 0.576 * | ||||||||||
| CBH | 0.455 | 0.793 ** | 0.778 ** | 0.592 * | 0.192 | 0.728 ** | 0.930 ** | |||||||||
| NAG | −0.030 | 0.602 * | 0.585 * | 0.550 * | 0.369 | 0.312 | 0.719 ** | 0.556 * | ||||||||
| PME | −0.481 | 0.542 * | 0.516 * | 0.260 | 0.549 * | −0.215 | 0.468 | 0.210 | 0.639 * | |||||||
| Total Reads | −0.292 | −0.026 | −0.005 | 0.217 | 0.277 | −0.219 | −0.154 | −0.313 | 0.003 | 0.178 | ||||||
| OTU Taxa | −0.528 * | −0.131 | −0.165 | 0.096 | 0.092 | −0.401 | −0.160 | −0.354 | 0.046 | 0.319 | 0.581 * | |||||
| Chao1 | −0.520 * | −0.171 | −0.199 | 0.059 | 0.032 | −0.393 | −0.197 | −0.377 | −0.002 | 0.293 | 0.547 * | 0.989 ** | ||||
| Pielou | −0.338 | −0.193 | −0.252 | −0.264 | −0.130 | −0.439 | −0.082 | −0.150 | −0.001 | 0.167 | −0.422 | 0.427 | 0.426 | |||
| Shannon | −0.512 | −0.223 | −0.281 | −0.148 | −0.053 | −0.519 * | −0.163 | −0.308 | 0.020 | 0.282 | −0.001 | 0.787 ** | 0.782 ** | 0.891 ** | ||
| Simpson | −0.584 * | −0.357 | −0.406 | −0.284 | −0.067 | −0.715 ** | −0.320 | −0.486 | −0.042 | 0.262 | 0.089 | 0.742 ** | 0.733 ** | 0.832 ** | 0.945 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Sun, H.; Yang, J.; Qin, J.; Shen, D.; Chen, Y. Variations in Soil Enzyme Activities and Microbial Communities along an Altitudinal Gradient on the Eastern Qinghai–Tibetan Plateau. Forests 2021, 12, 681. https://doi.org/10.3390/f12060681
Fan S, Sun H, Yang J, Qin J, Shen D, Chen Y. Variations in Soil Enzyme Activities and Microbial Communities along an Altitudinal Gradient on the Eastern Qinghai–Tibetan Plateau. Forests. 2021; 12(6):681. https://doi.org/10.3390/f12060681
Chicago/Turabian StyleFan, Shiyu, Hui Sun, Jiyuan Yang, Jihong Qin, Danjie Shen, and Yuexi Chen. 2021. "Variations in Soil Enzyme Activities and Microbial Communities along an Altitudinal Gradient on the Eastern Qinghai–Tibetan Plateau" Forests 12, no. 6: 681. https://doi.org/10.3390/f12060681
APA StyleFan, S., Sun, H., Yang, J., Qin, J., Shen, D., & Chen, Y. (2021). Variations in Soil Enzyme Activities and Microbial Communities along an Altitudinal Gradient on the Eastern Qinghai–Tibetan Plateau. Forests, 12(6), 681. https://doi.org/10.3390/f12060681






