Differing Responses to Cryphonectria parasitica at Two Indiana Locations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Information
2.2. Growth Measurements and Blight Evaluation
2.3. Cryphonectria parasitica Isolates
2.4. Isolate Growth and Attempted Conversion
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milgroom, M.G.; Wang, K.; Zhou, Y.; Lipari, S.E.; Kaneko, S. Intercontinental population structure of the chestnut blight fungus, Cryphonectria parasitica. Mycologia 1996, 88, 179–190. [Google Scholar] [CrossRef]
- Heiniger, U.; Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- Milgroom, M.G.; Cortesi, P. Biological control of chestnut blight with hypovirulence: A critical analysis. Annu. Rev. Phytopathol. 2004, 42, 311–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rea, G. What is blight resistance? J. Am. Chestnut Found. 2012, 26, 3. Available online: https://www.acf.org/wp-content/uploads/2016/08/Blight-Resistance.pdf (accessed on 14 April 2021).
- Anagnostakis, S. Biological control of chestnut blight. Science 1982, 215, 466–471. [Google Scholar] [CrossRef]
- Anagnostakis, S. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–27. [Google Scholar] [CrossRef]
- Hirsch, R. Blight resistance: It’s in the DNA. J. Am. Chestnut Found. 2012, 26, 20–22. Available online: http://www.rebeccahirsch.com/uploads/2/1/5/0/21500942/blight_resistance.pdf (accessed on 14 April 2021).
- Bragança, H.; Simões, S.; Onofre, N.; Santos, N. Factors influencing the incidence and spread of chestnut blight in northeastern Portugal. J. Plant Pathol. 2009, 91, 53–59. [Google Scholar] [CrossRef]
- Murolo, S.; Sciences, E.; Milvia, R. Phenotypic and molecular investigations on hypovirulent Cryphonectria parasitica in Italy. Plant Dis. 2018, 102, 540–545. [Google Scholar] [CrossRef] [Green Version]
- Karadžić, D.; Radulović, Z.; Sikora, K.; Stanivuković, Z.; Golubović Ćurguz, V.; Oszako, T.; Milenković, I. Characterisation and pathogenicity of Cryphonectria parasitica on sweet chestnut and sessile oak trees in Serbia. Plant Prot. Sci. 2019, 55, 191–201. [Google Scholar] [CrossRef]
- Dennert, F.; Rigling, M.; Meyer, J.B.; Schefer, C.; Augustiny, E.; Prospero, S. Testing the pathogenic potential of Cryphonectria parasitica and related species on three common European. Fagaceae. Front. Glob Chang. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Hepting, G.H. Death of the American chestnut. J. For. Hist. 1974, 18, 61–67. [Google Scholar] [CrossRef]
- Schlarbaum, S.E.; Hebard, F.; Spaine, P.C.; Kamalay, J.C. Three American tragedies: Chestnut blight, butternut canker, and Dutch elm disease. In Exotic Pests of Eastern Forests Conference Proceedings, Nashville, TN, USA, 8–10 April 1997; Britton, K.O., Ed.; U.S. Forest Service and Tennessee Exotic Pest Plant Council: Nashville, TN, USA, 1997; pp. 45–54. Available online: https://www.fs.usda.gov/treesearch/pubs/745 (accessed on 14 April 2021).
- Reynolds, D.L.; Burke, K.L. The effect of growth rate, age, and chestnut blight on American chestnut mortality. Castanea 2011, 76, 129–139. [Google Scholar] [CrossRef]
- Davelos, A.L.; Jarosz, A.M. Demography of American chestnut populations: Effects of a pathogen and a hyperparasite. J. Ecol. 2004, 92, 675–685. [Google Scholar] [CrossRef]
- Lee, S.H.; Moon, B.J.; Lee, J.K. Characteristics of hypovirulent strains of chestnut blight fungus, Cryphonectria parasitica, isolated in Korea. Mycobiology 2006, 34, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Prospero, S.; Rigling, D. Invasion genetics of the chestnut blight fungus Cryphonectria parasitica in Switzerland. Phytopathology 2012, 102, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Bryner, S.F.; Prospero, S.; Rigling, D. Dynamics of Cryphonectria hypovirus infection in chestnut blight cankers. Phytopathology 2014, 104, 918–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryner, S.F.; Sotirovski, K.; Akilli, S.; Risteski, M.; Perlerou, C.; Rigling, D. Informative value of canker morphology on the presence or absence of virus infection in chestnut blight cankers. For. Pathol. 2013, 43, 496–504. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, D. Plant health and quarantine regulations of the European Union for Cryphonectria parasitica. For. Snow Landsc. Res. 2001, 76, 402–404. Available online: https://www.dora.lib4ri.ch/wsl/islandora/object/wsl:15416 (accessed on 14 April 2021).
- Krstin, L.; Rigling, D.; Kraja, M. Chestnut blight fungus in Croatia: Diversity of vegetative compatibility types, mating types and genetic variability of associated Cryphonectria hypovirus 1. Plant Pathol. 2008, 57, 1086–1096. [Google Scholar] [CrossRef]
- Krstin, L.; Rigling, D. Diversity of vegetative compatibility types and mating types of Cryphonectria parasitica in Slovenia and occurrence of associated Cryphonectria hypovirus 1. Plant Pathol. 2012, 60, 752–761. [Google Scholar] [CrossRef]
- Zlatanov, T.; Velichkov, I.; Georgieva, M.; Hinkov, G.; Zlatanova, M.; Gogusev, G.; Eastaugh, C. Does management improve the state of chestnut (Castanea sativa L.) on Belasitsa Mountain, southwest Bulgaria? IForest 2015, 8, 860–865. [Google Scholar] [CrossRef] [Green Version]
- Krstin, L.; Katani, Z.; Ježi, M.; Poljak, I.; Nuskern, L.; Idžojti, M. Biological control of chestnut blight in Croatia: An interaction between host sweet chestnut, its pathogen Cryphonectria parasitica and the biocontrol agent Cryphonectria hypovirus 1. Pest. Manag. Sci. 2016, 73, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Prospero, S.; Cleary, M. Effects of Host Variability on the Spread of Invasive Forest Diseases. Forests 2017, 8, 80. [Google Scholar] [CrossRef] [Green Version]
- Lovat, C.-A.; Donnelly, D.J. Mechanisms and metabolomics of the host–pathogen interactions between Chestnut (Castanea species) and Chestnut blight (Cryphonectria parasitica). For. Pathol. 2019, 49, e12562. [Google Scholar] [CrossRef]
- García-Pedrajas, M.D.; Cañizares, M.C.; Sarmiento-Villamil, J.L.; Jaquat, A.G.; Dambolena, J.S. Mycoviruses in biological control: From basic research to field implementation. Phytopathology 2019, 109, 1828–1839. [Google Scholar] [CrossRef]
- Linder-Basso, D.; Dynek, J.N.; Hillman, B.I. Genome analysis of Cryphonectria hypovirus 4, the most common hypovirus species in North America. Virology 2005, 337, 192–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Jiang, D. New insights into mycoviruses and exploration for the biological control of crop fungal iseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef] [Green Version]
- Hillman, B.I.; Halpern, B.; Brown, M.P. A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology 1994, 201, 241–250. [Google Scholar] [CrossRef]
- Smart, C.D.; Yuan, W.; Foglia, R.; Nuss, D.L.; Fulbright, D.W.; Hillman, B.I. Cryphonectria hypovirus 3, a virus species in the family Hypoviridae with a single open reading frame. Virology 1999, 265, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Aghayeva, D.N.; Rigling, D.; Prospero, S. Low genetic diversity but frequent sexual reproduction of the chestnut blight fungus Cryphonectria parasitica in Azerbaijan. For. Pathol. 2017, 17, e12357. [Google Scholar] [CrossRef]
- Robin, C.; Capdevielle, X.; Martin, M.; Traver, C.; Colinas, C. Cryphonectria parasitica vegetative compatibility type analysis of populations in south-western France and northern Spain. Plant Pathol. 2009, 58, 527–535. [Google Scholar] [CrossRef]
- Adamčiková, K.; Onrušková, E.; Kádasi-Horáková, M.; Botu, M.; Kobza, M.; Achim, G. Distribution and population structure of the chestnut blight fungus in Romania. Plant Prot. Sci. 2015, 51, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Chira, D.; Bolea, V.; Chira, F.; Mantale, C.; Tăut, I.; Şimonca, V.; Diamandis, S. Biological control of Cryphonectria parasitica in Romanian protected sweet chestnut forests. Not. Bot. Horti Agrobot. Cluj Napoca 2017, 45, 632–638. [Google Scholar] [CrossRef] [Green Version]
- Krstin, L.; Katani, Z.; Repar, J.; Je, M.; Koba, A.; Mirna, Ć. Genetic diversity of Cryphonectria hypovirus 1, a biocontrol agent of chestnut blight, in Croatia and Slovenia. Microb. Ecol. 2020, 79, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Liu, Y.; Hillman, B.I.; Milgroom, M.G. Recombination and migration of Cryphonectria hypovirus 1 as inferred from gene genealogies and the coalescent. Genetics 2004, 166, 1611–1629. [Google Scholar] [CrossRef]
- Bryner, S.F.; Rigling, D.; Brunner, P.C. Invasion history and demographic pattern of Cryphonectria hypovirus 1 across European populations of the chestnut blight fungus. Ecol. Evol. 2012, 2, 3227–3241. [Google Scholar] [CrossRef]
- Risteski, B.M.; Milev, M.; Rigling, D.; Milgroom, M.G.; Bryner, S.F.; Sotirovski, K. Distribution of chestnut blight and diversity of Cryphonectria parasitica in chestnut forests in Bulgaria. Plant Pathol. 2013, 43, 437–443. [Google Scholar] [CrossRef]
- Ježić, M.; Mlinarec, J.; Vuković, R.; Katanić, Z.; Krstin, L.; Nuskern, L.; Poljak, I.; Idžojtić, M.; Tkalec, M.; Ćurković-Perica, M. Changes in Cryphonectria parasitica populations affect natural biological control of Chestnut Blight. Ecol. Epidemiol. 2018, 108, 870–877. [Google Scholar] [CrossRef] [Green Version]
- Mlinarec, J.; Je, M.; Cosi, J.; Curkovi, M. Multilocus PCR assay reveals high diversity of vegetative compatibility types in populations of Cryphonectria parasitica in Croatia. Plant Pathol. 2018, 67, 741–749. [Google Scholar] [CrossRef]
- Rigling, D.; Borst, N.; Cornejo, C.; Supatashvili, A.; Prospero, S. Genetic and phenotypic characterization of Cryphonectria hypovirus 1 from Eurasian Georgia. Viruses 2018, 10, 687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risteski, M.; Karin-Kujundžić, V.; Idžojtić, M.; Poljak, I.; Ćurković-Perica, M. Diversity of Cryphonectria parasitica in callused chestnut blight cankers on European and American chestnut. For. Pathol. 2019, 49, 1–8. [Google Scholar] [CrossRef]
- Zamora, B.P.; Martín, A.B.; Rigling, D.; Diez, J.J. Diversity of Cryphonectria parasitica in western Spain and identification of hypovirus-infected isolates. For. Pathol. 2012, 42, 412–419. [Google Scholar] [CrossRef]
- Kazmierczak, P.; Mccabe, P.; Turina, M.; Jacob-wilk, D.; van Alfen, N.K. The mycovirus CHV1 disrupts secretion of a developmentally regulated protein in Cryphonectria parasitica. J. Virol. 2012, 86, 6067–6074. [Google Scholar] [CrossRef] [Green Version]
- Çelİker, N.M.; Kaplan, C.; Onoğur, E.; Çetİnel, B.; Poyraz, D.; Uysal, A. Natural dissemination of hypovirulent Cryphonectria parasitica strain used for biological control of chestnut blight. Turk. J. Agric. For. 2017, 41, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Diamandis, B.S.; Perlerou, C.; Tziros, G.T.; Christopoulos, V.; Topalidou, E. Establishment and dissemination of hypovirulent strains of Cryphonectria parasitica in Greece. For. Pathol. 2015, 45, 408–414. [Google Scholar] [CrossRef]
- Trapiello, E.; Rigling, D.; González, A.J. Occurrence of hypovirus-infected Cryphonectria parasitica isolates in northern Spain: An encouraging situation for biological control of chestnut blight in Asturian forests. Eur. J. Plant Pathol. 2017, 149, 503–514. [Google Scholar] [CrossRef]
- Kolp, M.; Double, M.L.; Fulbright, D.W.; Macdonald, W.L.; Jarosz, A.M. Spatial and temporal dynamics of the fungal community of chestnut blight cankers on American chestnut (Castanea dentata) in Michigan and Wisconsin. Fungal Ecol. 2020, 45, 100925. [Google Scholar] [CrossRef]
- Double, M.L.; Nuss, D.L.; Rittenour, W.R.; Holásková, I.; Short, D.P.G.; Kasson, M.T.; MacDonald, W.L. Long-term field study of transgenic hypovirulent strains of Cryphonectria parasitica in a forest setting. For. Pathol. 2017, 47, 1–20. [Google Scholar] [CrossRef]
- Zamora, P.; González Casas, A.; Dueñas, M.; San Martin, R.; Diez, J.J. Factors influencing growth, sporulation and virus transfer in Cryphonectria parasitica isolates from Castilla and León (Spain). Eur. J. Plant Path. 2017, 148, 65–73. [Google Scholar] [CrossRef]
- Zamora, P.; Martín, A.; Dueñas, M.; Martín, R.S.; Diez, J.J. Cryphonectria parasitica isolates of the same vegetative compatibility type display different rates of transfer of CHV1 hypovirus. For. Pathol. 2015, 143, 767–777. [Google Scholar] [CrossRef]
- Akilli, S.; Katircioğlu, Y.Z.; Maden, S. Vegetative compatibility types of Cryphonectria parasitica, causal agent of chestnut blight, in the Black Sea region of Turkey. For. Pathol. 2009, 39, 390–396. [Google Scholar] [CrossRef]
- Daldal, M.; Erincik, Ö.; Wall, J.R. Geographical distribution of vegetative compatibility and mating types of Cryphonectria parasitica in İzmir, Manisa and Denizli provinces in western Turkey. For. Pathol. 2018, 48, 1–13. [Google Scholar] [CrossRef]
- Cornejo, C.; Rigling, D.; Kupper, Q.; Prospero, S. A multiplexed genotyping assay to determine vegetative incompatibility and mating type in Cryphonectria parasitica. Eur. J. Plant Pathol. 2019, 155, 81–91. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.; Romón-Ochoa, P.; Gorton, C.; Lewis, A.; Rees, H.; Van Der Linde, S.; Webber, J. High vegetative compatibility diversity of Cryphonectria parasitica infecting sweet chestnut (Castanea sativa) in Britain indicates multiple pathogen introductions. Plant Pathol. 2019, 68, 727–737. [Google Scholar] [CrossRef]
- Ko, Y.H.; So, K.K.; Chun, J.; Kim, D.H. Distinct roles of two DNA methyltransferases from Cryphonectria parasitica in fungal virulence, responses to hypovirus infection, and viral clearance. mBio 2021, 12, e02890–20. [Google Scholar] [CrossRef] [PubMed]
- Demené, A.; Laurent, B.; Cros-Arteil, S.; Boury, C.; Dutech, C. Chromosomal rearrangements but no change of genes and transposable elements repertoires in an invasive forest-pathogenic fungus. bioRxiv 2021, 3, e434572. [Google Scholar] [CrossRef]
- Oliva, J.; Castaño, C.; Baulenas, E.; Domínguez, G.; González-Olabarria, J.R.; Oliach, D. The impact of the socioeconomic environment on the implementation of control measures against an invasive forest pathogen. For. Ecol. Manag. 2016, 380, 118–127. [Google Scholar] [CrossRef]
- McKechnie, T.S. Some resistance genes against Cryphonectria parasitica may be strain-specific. In Proceedings of the 29th Southern Forest Tree Improvement Conference. Tree Improvement in North America: Past, Present, and Future, WFGA/SFTIC Joint Meeting, Galveston, TX, USA, 19–22 June 2007; p. 96. Available online: https://rngr.net/publications/tree-improvement-proceedings/sftic/2007 (Accessed on April 14, 2021).
- Sun, Q.; Choi, G.H.; Nuss, D.L. Hypovirus-responsive transcription factor gene pro1 of the chestnut blight fungus Cryphonectria parasitica is required for female fertility, asexual spore development, and stable maintenance of hypovirus Infection. Eukaryot. Cell 2009, 8, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Puia, C.E.; Grigorescu, A.; Miclea, R.V. The morphology and the biological control of Cryphonectria parasitica. Bull. UASVM Agric. 2012, 69, 353–355. Available online: http://www.researchgate.net/publication/268075980 (Accessed on April 14, 2021).
- Ugolini, F.; Massetti, L.; Pedrazzoli, F.; Tognetti, R.; Vecchione, A.; Zulini, L.; Maresi, G. Ecophysiological responses and vulnerability to other pathologies in European chestnut coppices, heavily infested by the Asian chestnut gall wasp. For. Ecol. Manag. 2014, 314, 38–49. [Google Scholar] [CrossRef]
- González-Varela, G.; González, A.J.; Milgroom, M.G. Clonal population structure and introductions of the chestnut blight fungus, Cryphonectria parasitica, in Asturias, northern Spain. Eur. J. Plant Pathol. 2011, 131, 67–79. [Google Scholar] [CrossRef]
- Tulowiecki, S.J. Modeling the historical distribution of American chestnut (Castanea dentata) for potential restoration in western New York State, US. For. Ecol. Manag. 2020, 462, e118003. [Google Scholar] [CrossRef]
- Griscom, H.; Griscom, B. Evaluating the ecological niche of American chestnut for optimal hybrid seedling reintroduction sites in the Appalachian ridge and valley province. New For. 2012, 43, 441–455. [Google Scholar] [CrossRef]
- Hoegger, P.J.; Heiniger, U.; Holdenrieder, O.; Rigling, D. Differential transfer and dissemination of hypovirus and nuclear and mitochondrial genomes of a hypovirus-infected Cryphonectria parasitica strain after introduction into a natural population. Appl. Environ. Microb. 2003, 69, 3767–3771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ježić, M.; Krstin, L.; Poljak, I.; Liber, Z.; Idžojtić, M.; Jelić, M.; Meštović, J.; Zebec, M.; Ćurković-Perica, M. Castanea sativa: Genotype-dependent recovery from chestnut blight. Tree Genet. Genomes 2014, 10, 101–110. [Google Scholar] [CrossRef]
- Bauman, J.M.; Howes, C.; Brian, K. Growth performance and chestnut blight incidence (Cryphonectria parasitica) of backcrossed chestnut seedlings in surface mine restoration. New For. 2014, 45, 813–828. [Google Scholar] [CrossRef]
- Newhouse, A.E.; Polin-mcguigan, L.D.; Baier, K.A.; Valletta, K.E.R.; Rottmann, W.H.; Tschaplinski, T.J.; Maynard, C.; Powell, W.A. Plant science transgenic American chestnuts show enhanced blight resistance and transmit the trait to T1 progeny. Plant Sci. 2014, 228, 88–97. [Google Scholar] [CrossRef]
- Stauder, C.M.; Nuss, D.L.; Zhang, D.; Double, M.L.; Macdonald, W.L.; Metheny, A.M.; Kasson, M.T. Enhanced hypovirus transmission by engineered super donor strains of the chestnut blight fungus, Cryphonectria parasitica, into a natural population of strains exhibiting diverse vegetative compatibility genotypes. Virology 2019, 528, 1–6. [Google Scholar] [CrossRef]
- Meyer, J.; Chalmandrier, L.; Fässler, F.; Schefer, C.; Rigling, D.; Prospero, S. Role of fresh dead wood in the epidemiology and the biological control of the chestnut blight fungus. Plant Dis. 2019, 103, 430–438. [Google Scholar] [CrossRef] [Green Version]
- Double, M.L.; Jarosz, A.M.; Fulbright, D.W.; Baines, A.D.; Macdonald, W.L. Evaluation of two decades of Cryphonectria parasitica hypovirus introduction in an American chestnut stand in Wisconsin. Phytopathology 2018, 108, 702–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bissegger, M.; Rigling, D.; Heiniger, U. Population Structure and Disease Development of Cryphonectria parasitica in European Chestnut Forests in the Presence of Natural Hypovirulence. Phytopathology 1997, 87, 50–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, B.F.S.; Holweg, C.L.; Rigling, D.; Metzler, B. Chestnut blight in south-western Germany: Multiple introductions of Cryphonectria parasitica and slow hypovirus spread. For. Pathol. 2012, 42, 397–404. [Google Scholar] [CrossRef]
- Akilli, S.; Ulbaş Serçe, C.; Zekai Katircioğlu, Y.; Maden, S.; Rigling, D. Characterization of hypovirulent isolates of the chestnut blight fungus, Cryphonectria parasitica from the Marmara and Black Sea regions of Turkey. Eur. J. Plant Pathol. 2013, 135, 323–334. [Google Scholar] [CrossRef] [Green Version]

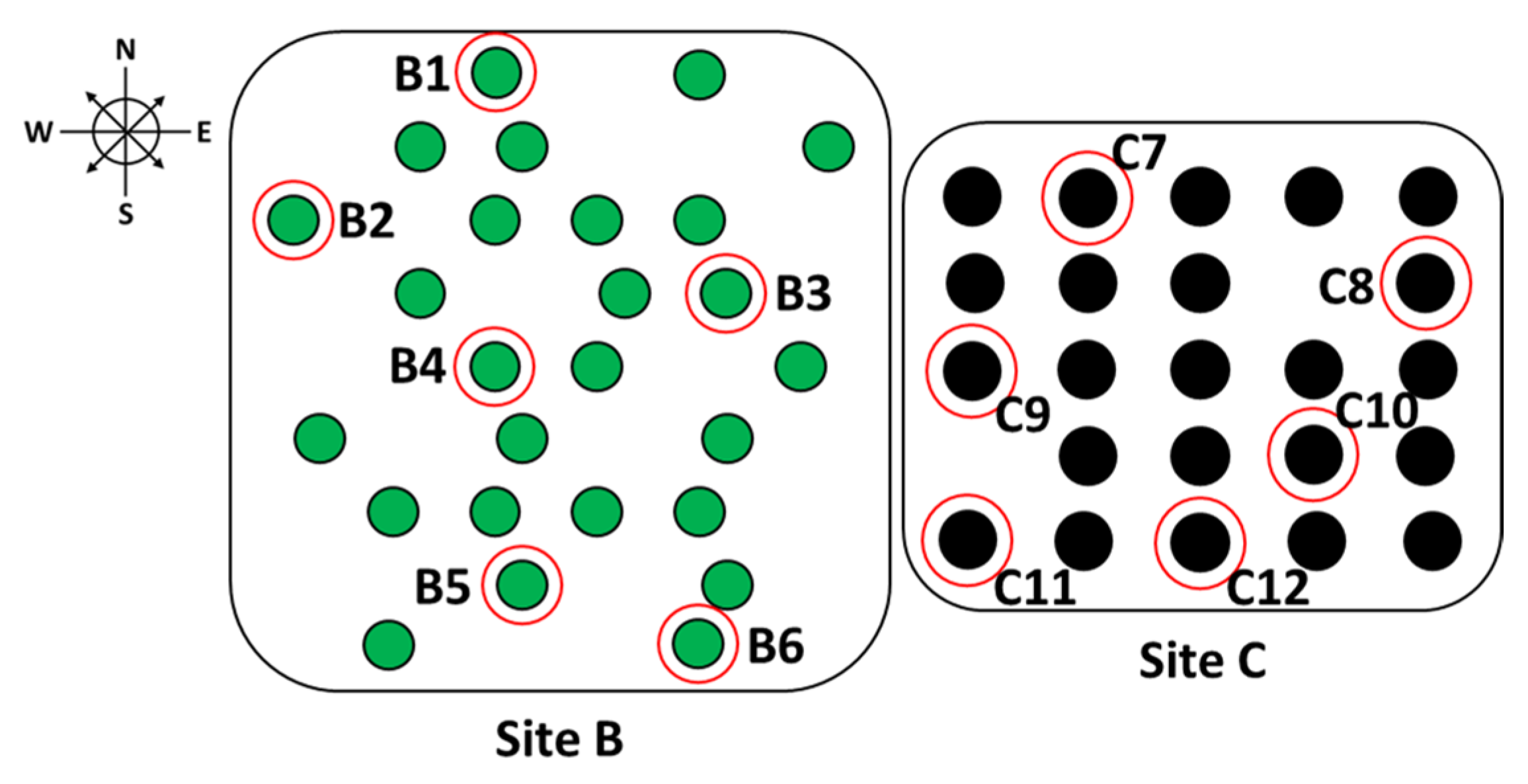
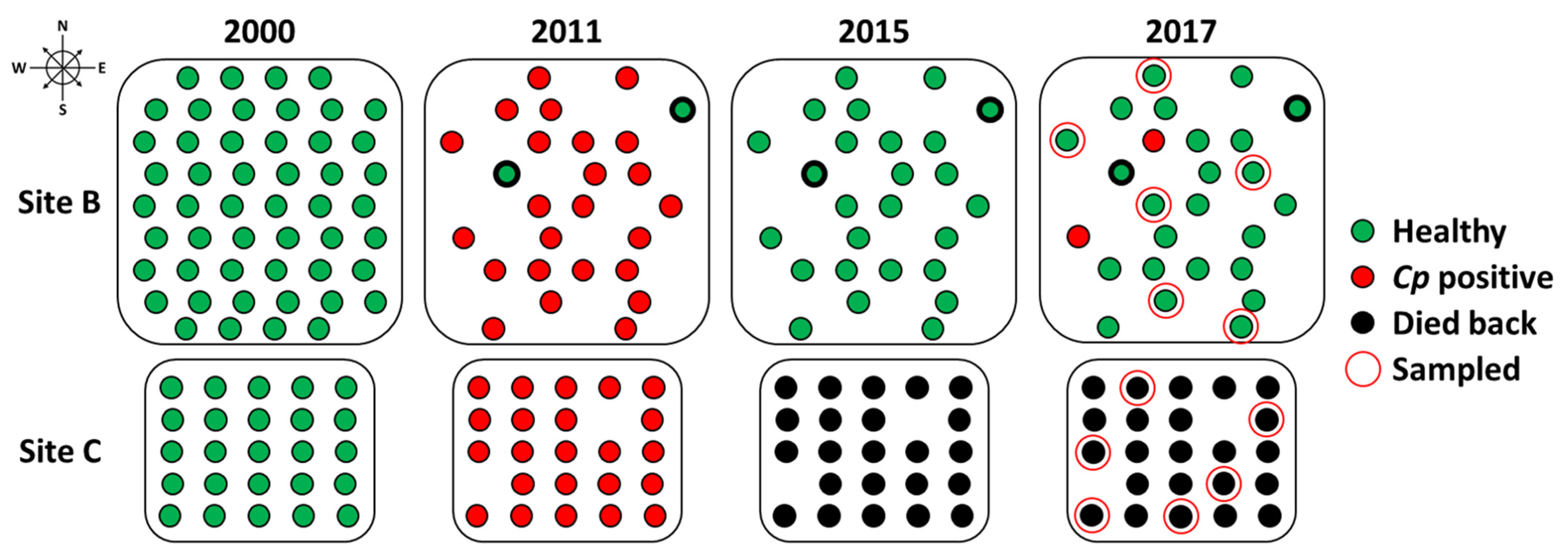

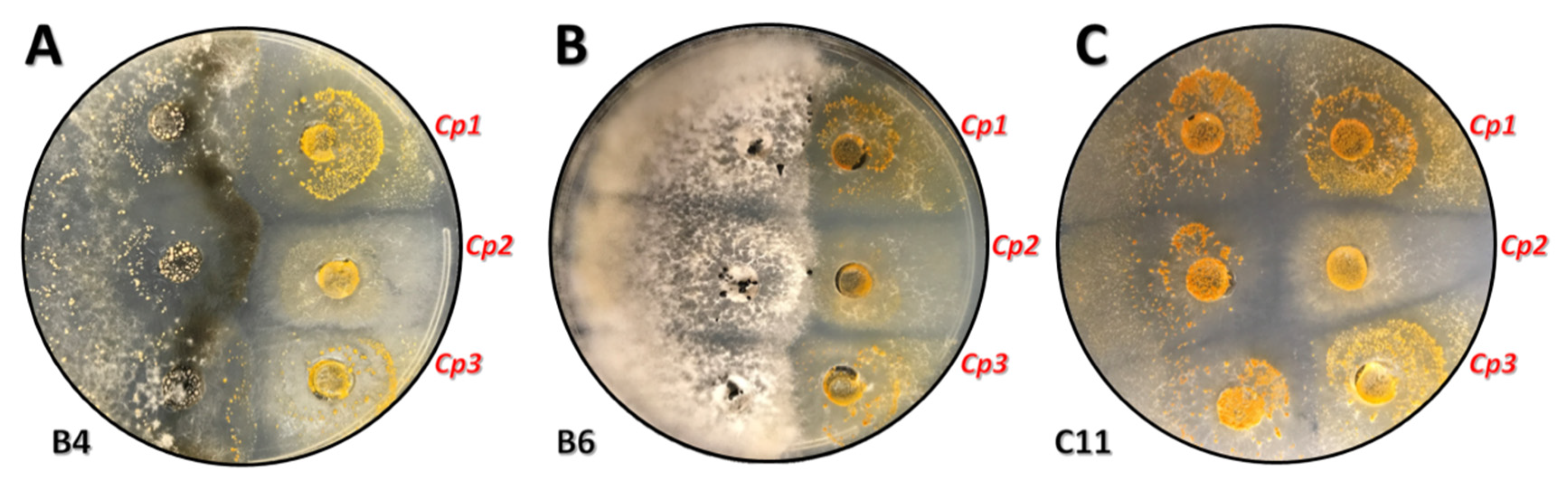

| Orchard | Avg. Height (ft) | Avg. DBH (in) | Avg. Canker Rating (1–3) | Avg. Disease Progress (%) |
|---|---|---|---|---|
| Site B | 17.5 | 3.2 | 1.6 | 92 |
| Site C | 22.1 | 5.1 | 1.5 | 100 |
| Orchard | Tree | No. Bark Samples (6 mm) | No. Fungal Isolates | Isolates per Sample (%) | Confirmed Cp Isolates |
|---|---|---|---|---|---|
| Site B | B1 | 9 | 6 | 67 | 0 |
| Site B | B2 | 10 | 2 | 20 | 0 |
| Site B | B3 | 9 | 1 | 11 | 0 |
| Site B | B4 | 12 | 7 | 58 | 0 |
| Site B | B5 | 14 | 1 | 7 | 0 |
| Site B | B6 | 12 | 4 | 33 | 0 |
| Site C | C7 | 8 | 3 | 38 | 2 |
| Site C | C8 | 5 | 2 | 40 | 2 |
| Site C | C9 | 7 | 4 | 57 | 0 |
| Site C | C10 | 6 | 3 | 50 | 1 |
| Site C | C11 | 7 | 4 | 57 | 3 |
| Site C | C12 | 8 | 2 | 25 | 1 |
| Totals | 107 | 39 | 39 * | 9 |
| Orchard | Tree | No. Bark Samples (6 mm) | No. Fungal Isolates | Isolates per Sample (%) | No. Putative CpHV Isolates |
|---|---|---|---|---|---|
| Site B | B2 | 14 | 6 | 43 | 0 |
| Site B | B5 | 12 | 3 | 25 | 0 |
| Site B | B6 | 8 | 2 | 25 | 0 |
| Site B | B7-y ‡ | 10 | 4 | 40 | 0 |
| Site B | B7-o † | 18 | 4 | 22 | 0 |
| Site B | B8-1 | 8 | 2 | 25 | 0 |
| Site B | B8-2 | 29 | 11 | 38 | 0 |
| Site B | B9-1 | 13 | 2 | 15 | 0 |
| Site B | B9-2 | 5 | 1 | 20 | 0 |
| Site B | B10 | 11 | 3 | 27 | 0 |
| Site B | B11 | 9 | 1 | 11 | 0 |
| Site B | B12 | 13 | 5 | 38 | 0 |
| Site B | B12-Cp | 19 | 6 | 32 | 1 |
| Site B | B13 | 13 | 5 | 38 | 0 |
| Site B | B13-Cp | 21 | 5 | 24 | 1 |
| Totals | 203 | 60 | 28 * | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lawson, S.S.; Ebrahimi, A.; McKenna, J.R. Differing Responses to Cryphonectria parasitica at Two Indiana Locations. Forests 2021, 12, 794. https://doi.org/10.3390/f12060794
Lawson SS, Ebrahimi A, McKenna JR. Differing Responses to Cryphonectria parasitica at Two Indiana Locations. Forests. 2021; 12(6):794. https://doi.org/10.3390/f12060794
Chicago/Turabian StyleLawson, Shaneka S., Aziz Ebrahimi, and James R. McKenna. 2021. "Differing Responses to Cryphonectria parasitica at Two Indiana Locations" Forests 12, no. 6: 794. https://doi.org/10.3390/f12060794
APA StyleLawson, S. S., Ebrahimi, A., & McKenna, J. R. (2021). Differing Responses to Cryphonectria parasitica at Two Indiana Locations. Forests, 12(6), 794. https://doi.org/10.3390/f12060794





