Environmental Heterogeneity Affecting Community Assembly Patterns and Phylogenetic Diversity of Three Forest Communities at Mt. Huangshan, China
Abstract
:1. Introduction
- Did the phylogenetic structure of three community assembly differ between tree and shrub plants, and how did phylogenetic diversity change in three vegetation regions?
- How did environmental variables (space, topography, and climate) influence and contribute to community diversity and community structure?
- Did the phylogenetic diversity of the three plant groups deviate faster or slower than predicted by the null model?
2. Materials and Methods
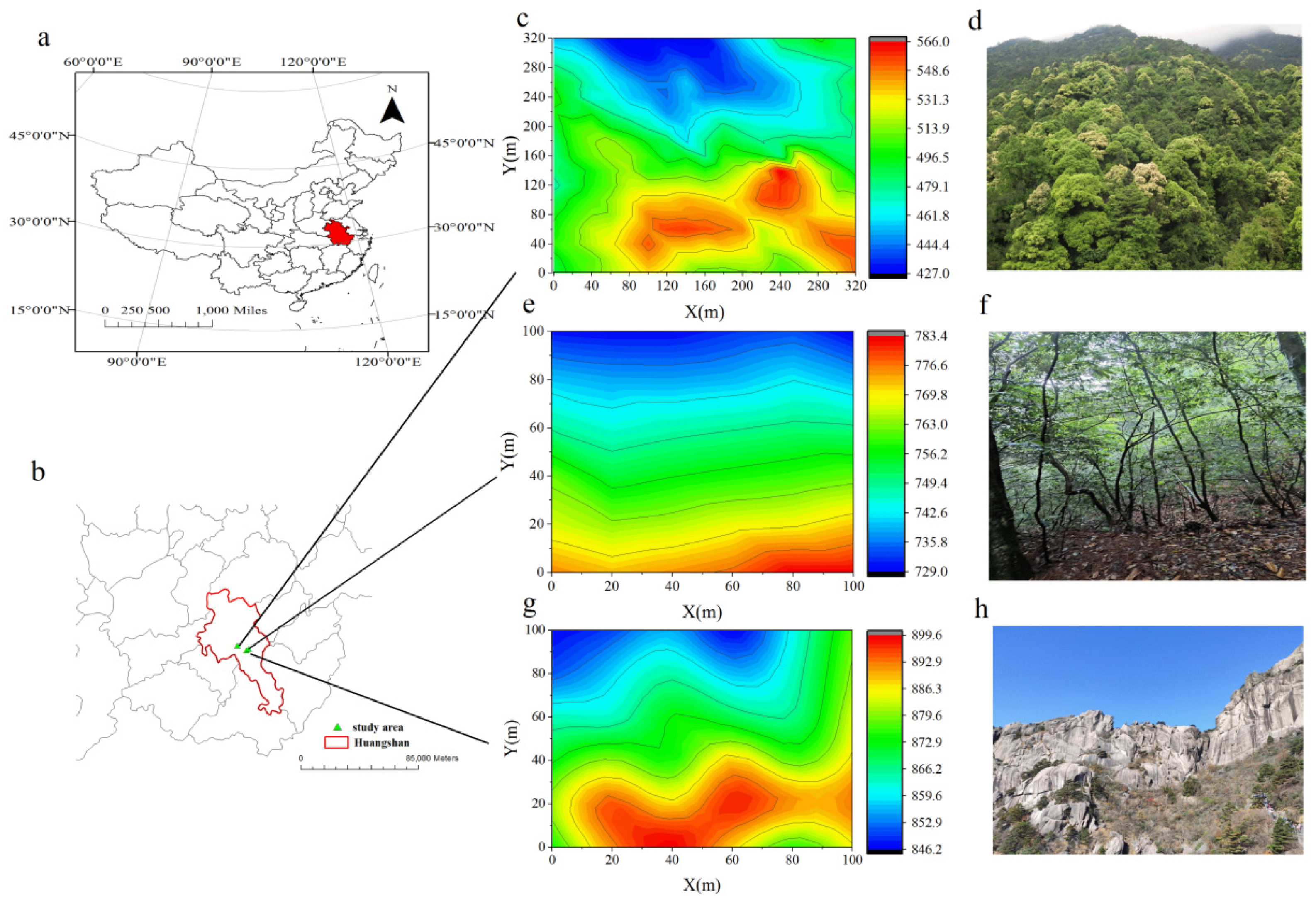
2.1. Study Area and Data Collection
2.2. Phylogenetic Tree Construction
2.3. Phylogenetic Community Structure
2.4. Phylogenetic α- and β-Diversity
2.5. Environmental Variables
2.6. Data Analysis
3. Results
3.1. DNA Amplification, Sequencing, and Phylogenetic Tree Construction
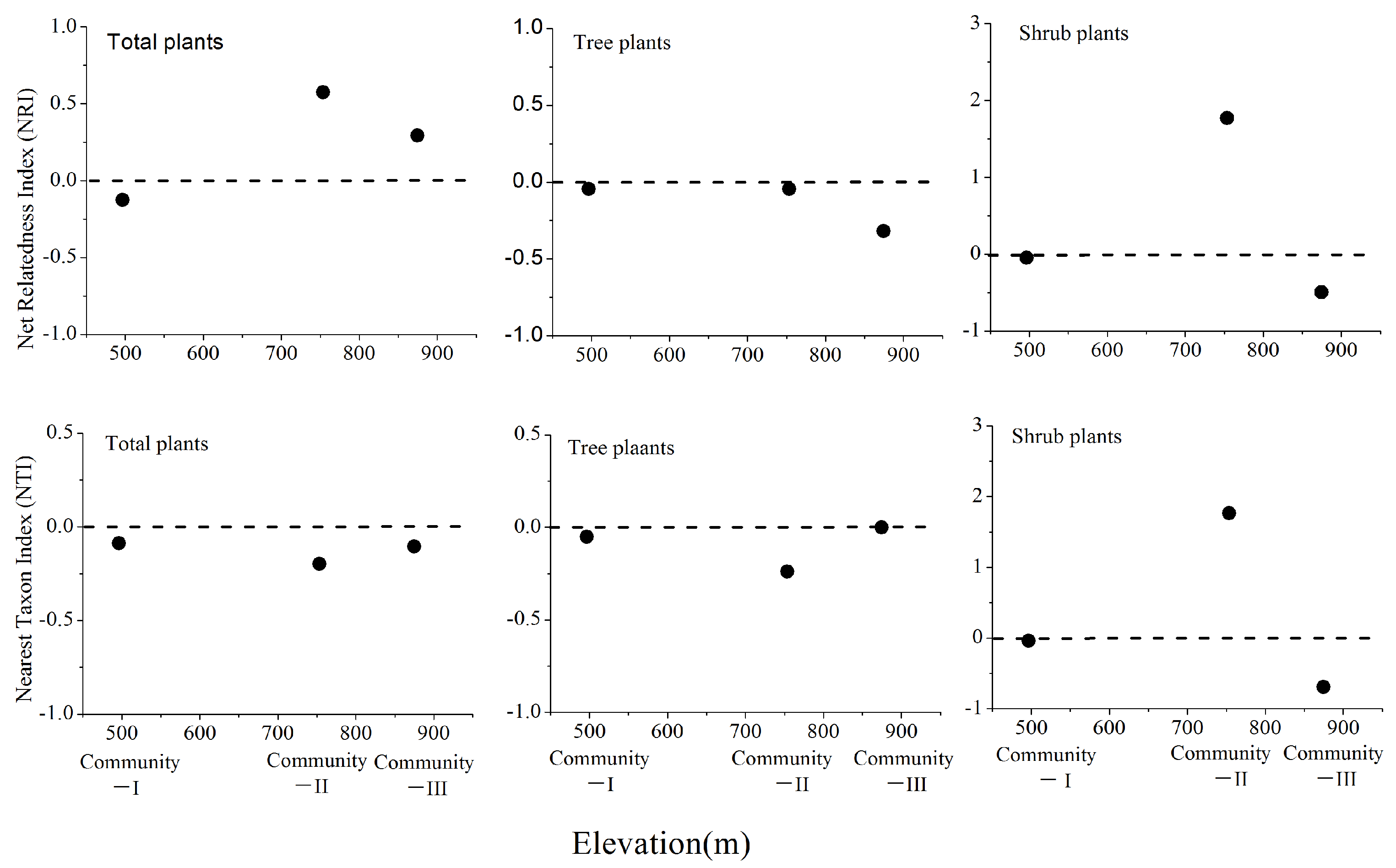
3.2. Phylogenetic Structure of Total, Tree, and Shrub Plants
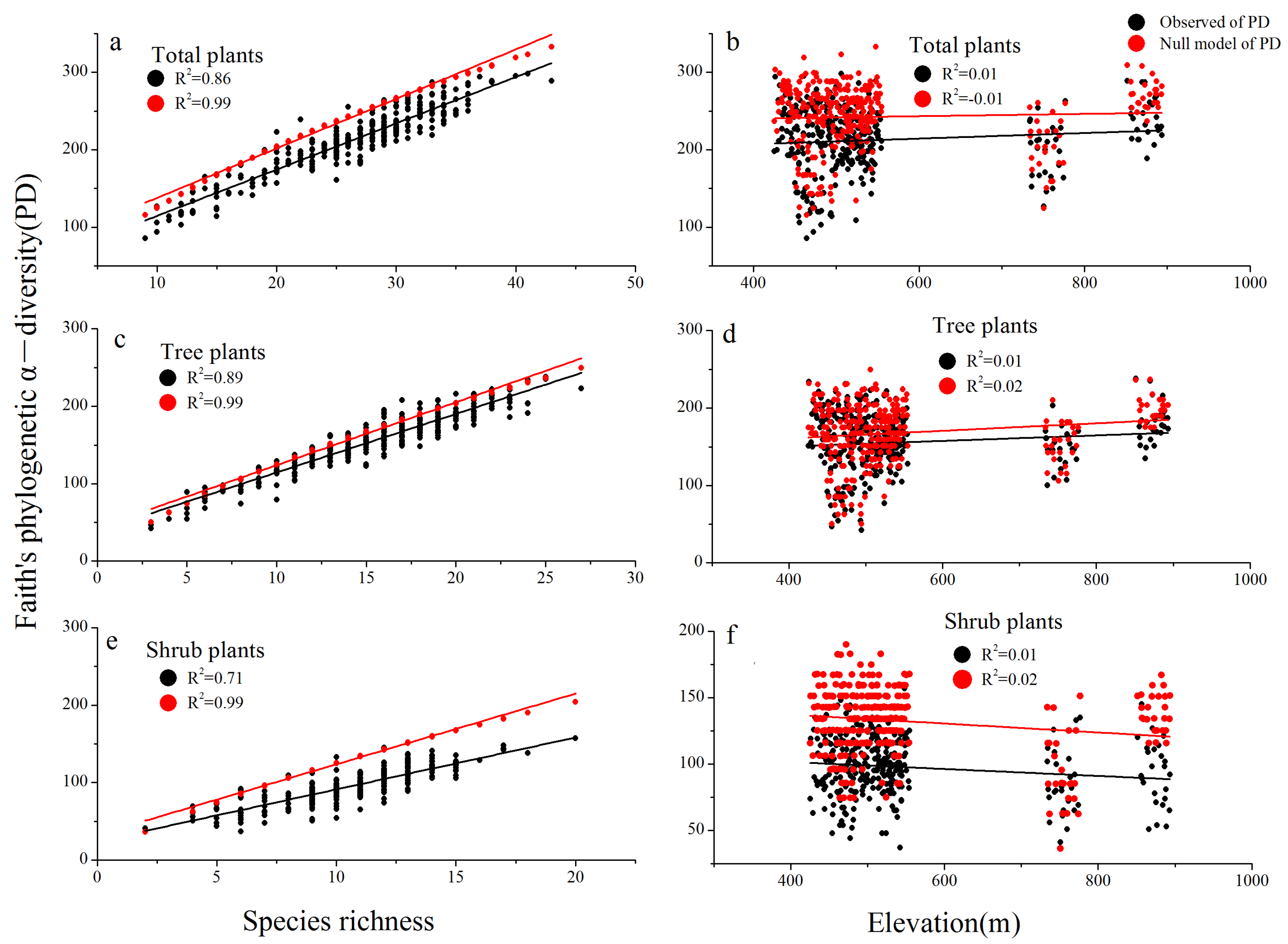
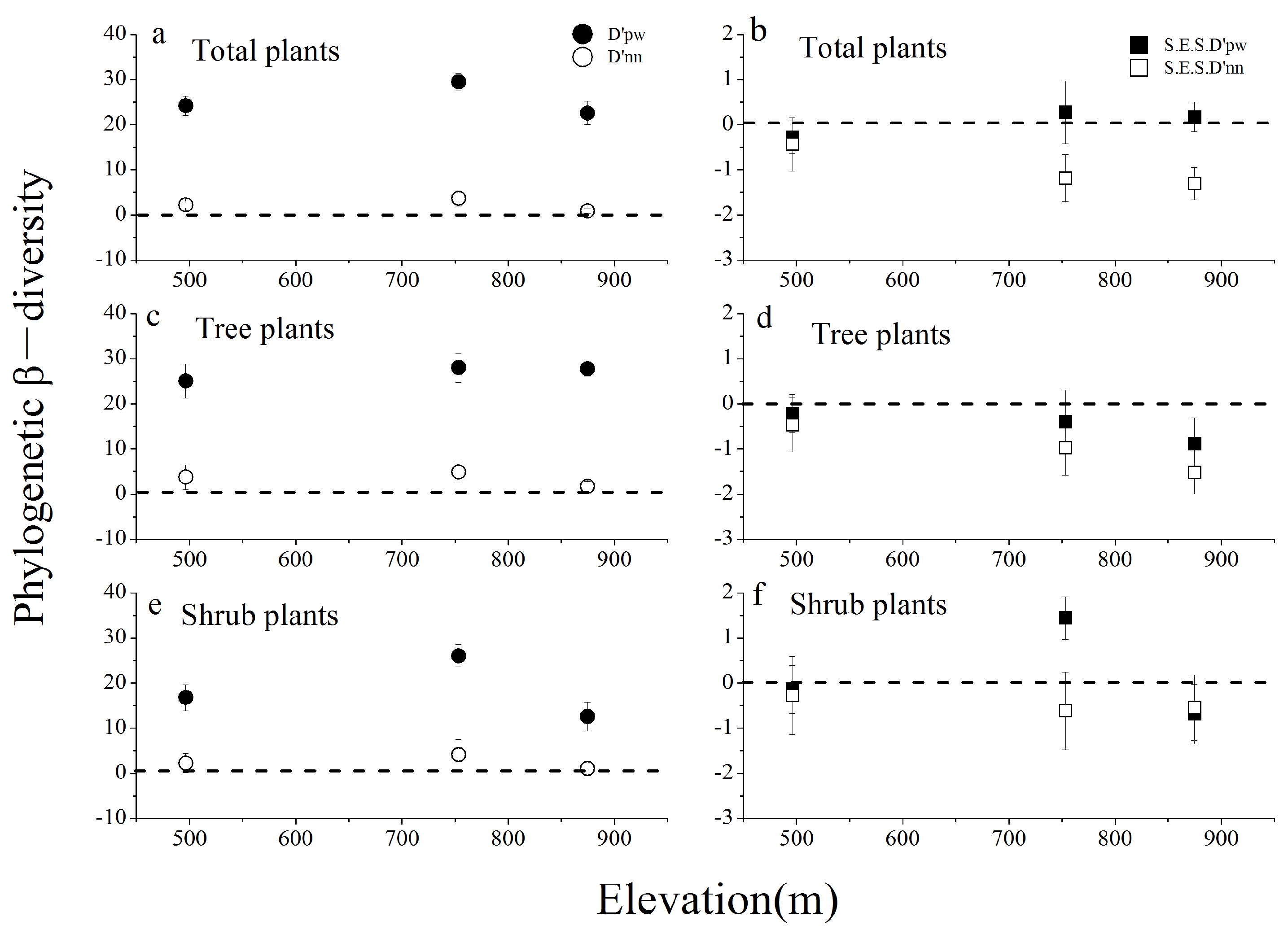
3.3. Species Richness and Phylogenetic Diversity across Three Different Communities
4. Discussion
4.1. Assessment of DNA Barcodes for Community Construction
4.2. Drivers of Phylogenetic Structure across Three Communities
4.3. Patterns and Determinants of Phylogenetic α- and β-Diversity in Three Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qian, H.; Hao, Z.; Zhang, J. Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J. Plant Ecol. 2014, 7, 154–165. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Sang, W.; Hausmann, A.; Axmacher, J.C. High phylogenetic diversity is preserved in species-poor high-elevation temperate moth assemblages. Sci. Rep. 2016, 6, 23045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavender-Bares, J.; Kozak, K.H.; Fine, P.; Kembel, S. The merging of community ecology and phylogenetic biology. Ecol. Lett. 2009, 12, 693–715. [Google Scholar] [CrossRef]
- Tilman, D. Niche tradeoffs, neutrality, and community structure: A stochastic theory of resource competition, invasion, and community assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 10854–10861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.Y.; Huang, W.Q.; Liu, M. Construction of forest biodiversity monitoring network based on continuous forest inven-tory system: A case study in Hunan Province. For. Resour. Manag. 2012, 0, 51–58. [Google Scholar]
- Chun, J.-H.; Lee, C.-B. Diversity patterns and phylogenetic structure of vascular plants along elevational gradients in a mountain ecosystem, South Korea. J. Mt. Sci. 2018, 15, 280–295. [Google Scholar] [CrossRef]
- Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography; Princeton University Press: Princeton, NJ, USA, 2001. [Google Scholar]
- Zheng, Y.; Dong, L.; Li, Z.; Zhang, J.; Li, Z.; Miao, B.; Jia, C.; Liang, C.; Wang, L.; Li, F.Y. Phylogenetic structure and formation mechanism of shrub communities in arid and semiarid areas of the Mongolian Plateau. Ecol. Evol. 2019, 9, 13320–13331. [Google Scholar] [CrossRef]
- Zu, K.; Luo, A.; Shrestha, N.; Liu, B.; Wang, Z.; Zhu, X. Altitudinal biodiversity patterns of seed plants along Gongga Mountain in the southeastern Qinghai–Tibetan Plateau. Ecol. Evol. 2019, 9, 9586–9596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, K.C.; Liu, Y.N.; Shen, Z.H.; He, F.; Fang, J. Community assembly: The relative importance of neutral theory and niche theory. Biodivers. Sci. 2009, 17, 579–593. [Google Scholar]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and Community Ecology. Annu. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef] [Green Version]
- Cao, K.; Rao, M.D.; Yu, J.Z.; Liu, X.; Chen, J. The phylogenetic signal of functional traits and their effects on community structure in an evergreen broad-leaved forest. Biodivers. Sci. 2013, 21, 564–571. [Google Scholar] [CrossRef]
- Lu, M.M.; Huang, X.C.; Ci, X.Q.; Yang, G.P.; Li, J. Phylogenetic community structure of subtropical forests along elevational gradients in Ailao Mountains of southwest China. Biodivers. Sci. 2014, 22, 438. [Google Scholar] [CrossRef]
- Bryant, J.A.; Lamanna, C.; Morlon, H.; Kerkhoff, A.J.; Enquist, B.; Green, J.L. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. USA 2008, 105, 11505–11511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, C.H.; Parra, J.L.; Rahbek, C.; McGuire, J.A. Phylogenetic structure in tropical hummingbird communities. Proc. Natl. Acad. Sci. USA 2009, 106, 19673–19678. [Google Scholar] [CrossRef] [Green Version]
- Yakimov, B.N.; Gerasimova, A.S.; Zhang, S.; Ma, K.; Zhang, Y. Phylogenetic α- and β-diversity elevational gradients reveal consistent patterns of temperate forest community structure. Acta Oecol. 2020, 109, 103657. [Google Scholar] [CrossRef]
- Faith, D.P. Phylogenetic Diversity and Conservation Evaluation: Perspectives on Multiple Values, Indices, and Scales of Application. In Phylogenetic Diversit: Applications and Challenges in Biodiversity Science; Scherson, R., Faith, D., Eds.; Springer: Cham, Switzerland; Berlin, Germany, 2018; pp. 1–26. [Google Scholar] [CrossRef]
- FitzJohn, R.G.; Pennell, M.W.; Zanne, A.E.; Stevens, P.F.; Tank, D.C.; Cornwell, W.K. How much of the world is woody? J. Ecol. 2014, 102, 1266–1272. [Google Scholar] [CrossRef]
- Feng, G.; Mi, X.C.; Bøcher, P.K.; Mao, L.F.; Sandel, B.; Cao, M.; Ye, W.H.; Hao, Z.Q.; Gong, H.D.; Zhang, Y.T.; et al. Relative roles of local disturbance, current climate and paleoclimate in determining phylogenetic and functional diversity in Chinese forests. Biogeosciences 2014, 11, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Che, Y.; Jiao, J.; Li, L.; Jiang, X. Exploring the community phylogenetic structure along the slope aspect of subalpine meadows in the eastern Qinghai-Tibetan Plateau, China. Ecol. Evol. 2019, 9, 5270–5280. [Google Scholar] [CrossRef] [PubMed]
- González-Caro, S.; Umaña, M.N.; Alvarez-Davila, E.; Stevenson, P.R.; Swenson, N.G. Phylogenetic alpha and beta diversity in tropical tree assemblages along regional-scale environmental gradients in northwest South America. J. Plant Ecol. 2014, 7, 145–153. [Google Scholar] [CrossRef]
- Kim, H.; Chun, J.; Lee, C. Plant Diversity and Phylogenetic Community Structure along Environmental Gradients in a Temperate Forest, South Korea. J. Anim. Plant Sci. 2020, 30, 958. [Google Scholar] [CrossRef] [Green Version]
- David, R.; Bomhard, B. Mapping Direct Human Influence on the World’s Mountain Areas. Mt. Res. Dev. 2012, 32, 197–202. [Google Scholar] [CrossRef]
- Culmsee, H.; Leuschner, C. Consistent patterns of elevational change in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr. 2013, 40, 1997–2010. [Google Scholar] [CrossRef]
- Letts, M.G.; Mulligan, M.; Rincon-Romero, M.E.; Bruijnzeelet, L.A. Environmental Controls on Photosynthetic Rates of Lower Montane Cloud Forest Vegetation in South-Western Colombia; Cambridge University Press: Cambridge, UK, 2010; pp. 465–478. [Google Scholar]
- Vamosi, S.M.; Queenborough, S.A. Breeding systems and phylogenetic diversity of seed plants along a large-scale elevational gradient. J. Biogeogr. 2010, 37, 465–476. [Google Scholar] [CrossRef]
- Rahbek, C. The role of spatial scale and the perception of large-scale species-richness patterns. Ecol. Lett. 2004, 8, 224–239. [Google Scholar] [CrossRef]
- Ding, H.; Fang, Y.; Yang, X.; Yuan, F.; He, L.; Yao, J.; Wu, J.; Chi, B.; Li, Y.; Chen, S.; et al. Community characteristics of a subtropical evergreen broad-leaved forest in Huangshan, Anhui Province, East China. Biodivers. Sci. 2016, 24, 875–887. [Google Scholar] [CrossRef]
- Yuan, J.F.; Hu, R.Y.; Shen, J.H.; Zhang, L.; Zhang, X.Y.; Yu, M.J. Comparison of species composition and diversity of four successional forest communities in Zhejiang Province, East China. Bull. Bot. Res. 2011, 31, 61–66. [Google Scholar] [CrossRef]
- Li, D.; Du, Y.; Xu, W.; Peng, D.; Primack, R.; Chen, G.; Mao, L.F.; Ma, K. Phylogenetic conservatism of fruit development time in Chinese angiosperms and the phylogenetic and climatic correlates. Glob. Ecol. Conserv. 2021, 27, e01543. [Google Scholar] [CrossRef]
- Ma, K.P. Large scale permanent plots: Important platform for long term research on biodiversity in forest ecosystem. Chin. J. Plant Ecol. 2008, 32, 237. [Google Scholar]
- Forest, F.; Grenyer, R.; Rouget, M.; Davies, T.J.; Cowling, R.M.; Faith, D.P.; Balmford, A.; Manning, J.C.; Procheş, Ş.; Van Der Bank, M.; et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature 2007, 445, 757–760. [Google Scholar] [CrossRef]
- Sargent, R.D.; Ackerly, D. Plant–pollinator interactions and the assembly of plant communities. Trends Ecol. Evol. 2008, 23, 123–130. [Google Scholar] [CrossRef] [PubMed]
- APG-Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and fam-ilies of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Pashirzad, M.; Ejtehadi, H.; Vaezi, J.; Shefferson, R.P. Spatial scale-dependent phylogenetic signal in species distributions along geographic and elevation gradients in a mountainous rangeland. Ecol. Evol. 2018, 8, 10364–10373. [Google Scholar] [CrossRef] [PubMed]
- Joly, S.; Davies, T.J.; Archambault, A.; Bruneau, A.; Derry, A.; Kembel, S.W.; Peres-Neto, P.; Vamosi, J.; Wheeler, T.A. Ecology in the age of DNA barcoding: The resource, the promise and the challenges ahead. Mol. Ecol. Resour. 2013, 14, 221–232. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, N.C.; Chen, B.F. DNA barcoding of life: A classification of uses according to function and scale after ten years of development. Biodivers. Sci. 2013, 21, 616–627. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Swenson, N.G.; Enquist, B.; Thompson, J.; Zimmerman, J.K. The Influence of Spatial and Size Scale on Phylogenetic Relatedness in Tropical Forest Communities. Ecology 2007, 88, 1770–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraft, N.J.B.; Cornwell, W.; Webb, C.; Ackerly, D. Trait Evolution, Community Assembly, and the Phylogenetic Structure of Ecological Communities. Am. Nat. 2007, 170, 271–283. [Google Scholar] [CrossRef]
- Chai, Y.F.; Yue, M. Research advances in plant community assembly mechanisms. Acta Ecol. Sin. 2016, 36, 4557–4572. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Entsminger, G.L. Swap algorithms in null model analysis. Ecology 2003, 84, 532–535. [Google Scholar] [CrossRef]
- Luo, Y.H. Community Assembly and Turnover Mechanisms Subalpine Forests along Elevational Gradient in Yulong Mountains in Northwest Yunnan, China; Yunnan University: Kunming, China, 2016. [Google Scholar]
- Dray, S.; Legendre, P.; Neto, P.P. Spatial modelling: A comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol. Model. 2006, 196, 483–493. [Google Scholar] [CrossRef]
- Legendre, P.; Mi, X.; Ren, H.; Ma, K.; Yu, M.; Sun, I.-F.; He, F. Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 2009, 90, 663–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichstein, J.W. Multiple regression on distance matrices: A multivariate spatial analysis tool. Plant Ecol. 2007, 188, 117–131. [Google Scholar] [CrossRef]
- Potter, K.M. From Genes to Ecosystems: Measuring Evolutionary Diversity and Community Structure with Forest Inventory and Analysis (FIA) Data. USDA For. Serv. Proc. 2009, 56, 49–64. Available online: https://www.researchgate.net/publication/228509453 (accessed on 10 January 2009).
- Mayfield, M.M.; Levine, J.M. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 2010, 13, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-H.; Zhu, X.-X.; Niu, Y.; Sun, H. Phylogenetic clustering and overdispersion for alpine plants along elevational gradient in the Hengduan Mountains Region, southwest China. J. Syst. Evol. 2014, 52, 280–288. [Google Scholar] [CrossRef]
- Gastauer, M.; Saporetti-Junior, A.W.; Valladares, F.; Meira-Neto, J.A.A. Phylogenetic community structure reveals differences in plant community assembly of an oligotrophic white-sand ecosystem from the Brazilian Atlantic Forest. Acta Bot. Bras. 2017, 31, 531–538. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chen, S.; Hu, G.; Mwachala, G.; Yan, X.; Wang, Q. Species richness and phylogenetic diversity of seed plants across vegetation zones of Mount Kenya, East Africa. Ecol. Evol. 2018, 8, 8930–8939. [Google Scholar] [CrossRef]
- Huang, J.X.; Ye, W.H.; Lian, J.Y.; Cao, H.L. Detecting the influence of phylogenetic structure, environmental factors and PCNM factors in population dynamics in a subtropical forest community in Guangdong, China. Chin. Sci. Bull. 2014, 59, 3471–3478. [Google Scholar] [CrossRef]
- Yuan, Z.L.; Chen, Y.; Wei, B.L.; Zhang, B.Q.; Wang, D.Y.; Ye, Y.Z. Species habitat correlation analysis in temper-ate-subtropical ecological transition zone. Acta Ecol. Sin. 2013, 33, 7819–7826. [Google Scholar]
- Xiao, Y.; Yang, L.; Nie, X.; Li, C.; Xiong, F.; Wang, L.; Zhou, G. Examining differences in phylogenetic composition enhances understanding of the phylogenetic structure of the shrub community in the northeastern Qinghai-Tibetan Plateau. Ecol. Evol. 2020, 10, 6723–6731. [Google Scholar] [CrossRef]
- Arponen, A. Prioritizing species for conservation planning. Biodivers. Conserv. 2012, 21, 875–893. [Google Scholar] [CrossRef]
- Yang, J.; Swenson, N.G.; Zhang, G.; Ci, X.; Cao, M.; Sha, L.; Li, J.; Slik, J.W.F.; Lin, L. Local-scale Partitioning of Functional and Phylogenetic Beta Diversity in a Tropical Tree Assemblage. Sci. Rep. 2015, 5, 12731. [Google Scholar] [CrossRef]
- Wei, W.; Luo, Z.R.; Zhou, R.F.; Wang, W.; Luo, Z.R.; Zhou, R.F. Habitat associations of woody plant species in Baishanzu subtropical broad-leaved evergreen forest. Biodivers. Sci. 2011, 19, 134–142. [Google Scholar] [CrossRef]
| Methods | Support Values (%) | |||
|---|---|---|---|---|
| Strong Ratchet (≥85%) | Moderate Ratchet (>75%–85%) | Weak Ratchet (>50%–75%) | Weak Ratchet (<50%) | |
| APGIV | - | - | - | - |
| Barcode tree without constraint | 59.88(110) | 13.77(23) | 15.57(26) | 10.78(18) |
| Barcode tree with constraint | 78.23(134) | 4.7(10) | 11.76(20) | 5.29(9) |
| Factor | Group | Variables | AdjR2Cum | F | p |
|---|---|---|---|---|---|
| Space | Total | PCNM1 | 0.21 | 81.56 | 0.001 |
| PCNM3 | 0.22 | 6.21 | 0.012 | ||
| PCNM30 | 0.24 | 5.25 | 0.024 | ||
| PCNM28 | 0.25 | 4.29 | 0.034 | ||
| PCNM16 | 0.26 | 4.24 | 0.038 | ||
| Trees | PCNM15 | 0.02 | 6.83 | 0.0071 | |
| PCNM29 | 0.03 | 5.39 | 0.0189 | ||
| PCNM4 | 0.05 | 5.01 | 0.0261 | ||
| PCNM27 | 0.06 | 4.11 | 0.0423 | ||
| PCNM13 | 0.06 | 4.02 | 0.0413 | ||
| Shrubs | PCNM3 | 0.36 | 178.87 | 0.001 | |
| PCNM1 | 0.48 | 66.37 | 0.001 | ||
| PCNM4 | 0.49 | 10.16 | 0.004 | ||
| PCNM11 | 0.50 | 8.02 | 0.013 | ||
| PCNM8 | 0.51 | 8.07 | 0.006 | ||
| PCNM5 | 0.52 | 5.96 | 0.017 | ||
| Climate | Total | Bio 15 | 0.21 | 78.59 | 0.0001 |
| Bio 10 | 0.26 | 22.69 | 0.0001 | ||
| Trees | Bio 14 | 0.02 | 7.31 | 0.0077 | |
| Shrubs | Bio 1 | 0.49 | 303.69 | 0.0001 | |
| Topography | Total | Elevation | 0.17 | 63.10 | 0.0004 |
| Aspect | 0.20 | 11.19 | 0.0008 | ||
| Trees | Elevation | 0.01 | 4.18 | 0.043 | |
| Shrubs | Aspect | 0.11 | 36.48 | 0.0004 | |
| Elevation | 0.12 | 7.43 | 0.0078 |
| Total Plants | Tree Plants | Shrub Plants | |||||
|---|---|---|---|---|---|---|---|
| R2 | p | R2 | p | R2 | p | ||
| D’pw | Space | 0.379 | 0.001 | 0.291 | 0.001 | 0.319 | 0.001 |
| Topography | 0.094 | 0.001 | 0.175 | 0.001 | 0.056 | 0.001 | |
| Climate | 0.549 | 0.001 | 0.250 | 0.001 | 0.536 | 0.001 | |
| Space + Topography + Climate | 0.629 | 0.001 | 0.311 | 0.001 | 0.624 | 0.001 | |
| D’nn | Space | 0.453 | 0.001 | 0.418 | 0.001 | 0.413 | 0.001 |
| Topography | 0.204 | 0.001 | 0.275 | 0.001 | 0.149 | 0.001 | |
| Climate | 0.647 | 0.001 | 0.466 | 0.001 | 0.650 | 0.001 | |
| Space + Topography + Climate | 0.672 | 0.001 | 0.490 | 0.001 | 0.677 | 0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, T.; Wang, N.; Xie, L.; Chen, S.; Zhao, R.; Feng, Y.; Li, Y.; Ding, H.; Fang, Y. Environmental Heterogeneity Affecting Community Assembly Patterns and Phylogenetic Diversity of Three Forest Communities at Mt. Huangshan, China. Forests 2022, 13, 133. https://doi.org/10.3390/f13010133
Lv T, Wang N, Xie L, Chen S, Zhao R, Feng Y, Li Y, Ding H, Fang Y. Environmental Heterogeneity Affecting Community Assembly Patterns and Phylogenetic Diversity of Three Forest Communities at Mt. Huangshan, China. Forests. 2022; 13(1):133. https://doi.org/10.3390/f13010133
Chicago/Turabian StyleLv, Ting, Ningjie Wang, Lei Xie, Shuifei Chen, Rong Zhao, Yueyao Feng, Yao Li, Hui Ding, and Yanming Fang. 2022. "Environmental Heterogeneity Affecting Community Assembly Patterns and Phylogenetic Diversity of Three Forest Communities at Mt. Huangshan, China" Forests 13, no. 1: 133. https://doi.org/10.3390/f13010133
APA StyleLv, T., Wang, N., Xie, L., Chen, S., Zhao, R., Feng, Y., Li, Y., Ding, H., & Fang, Y. (2022). Environmental Heterogeneity Affecting Community Assembly Patterns and Phylogenetic Diversity of Three Forest Communities at Mt. Huangshan, China. Forests, 13(1), 133. https://doi.org/10.3390/f13010133






