Variation Pattern and Genome-Wide Association Study of Leaf Phenotypic Traits among Ancient Ginkgo biloba L. Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Information
2.2. Measurement of Leaf Traits
2.3. Library Construction and Sequencing
2.4. SNP Calling and Filtering
2.5. GWASs and Associated Gene Detection
2.6. Statistical Analysis
3. Results
3.1. Variations in Leaf Traits among Populations
3.2. Variations in Leaf Traits within Populations
3.3. Correlations between Leaf Traits and Climatic Factors
3.4. Genotyping by Sequencing
3.5. SNP Calling
3.6. Genome-Wide Association Study
3.7. Genes Related to Leaf Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, Z.Y.; Zheng, S.L. Palaeobiology: The missing link in Ginkgo evolution. Nature 2003, 423, 821–822. [Google Scholar] [CrossRef]
- Xiang, Y.H.; Xiang, B.X.; Zhao, M.S.; Wang, Z.L. A report on the natural forest with Ginkgo population in west Tianmu moutains, Zhejiang province. Guizhou Sci. 2000, 18, 77–92. [Google Scholar]
- Fan, X.X.; Shen, L.; Zhang, X.; Chen, X.Y.; Fu, C.X. Assessing genetic diversity of Ginkgo biloba L. (Ginkgoaceae) populations from China by RAPD markers. Biochem. Genet. 2004, 42, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zeng, Z.; Chen, Y.Y.; Chen, C.; Qiu, Y.X.; Fu, C.X. Glacial refugia of Ginkgo biloba and human impact on its genetic diversity: Evidence from chloroplast DNA. J. Integr. Plant Biol. 2008, 50, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.Q.; Yang, Y.C.; Ohsawa, M.; Yi, S.R.; Momohar, A.; Su, W.H.; Wang, H.C.; Zhang, Z.Y.; Peng, M.C.; Wu, Z.L. Evidence for the persistence of wild Ginkgo biloba (Ginkgoaceae) populations in the Dalou Mountains, southwestern China. Am. J. Bot. 2012, 99, 1408–1414. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Mu, K.M.; Xu, M.; Ma, X.Y.; Ni, Z.X.; Wang, J.W.; Xu, L.A. Variation in the concentrations of major secondary metabolites in Ginkgo leaves from different geographical populations. Forests 2017, 8, 266. [Google Scholar] [CrossRef] [Green Version]
- Gattmann, M.; McAdam, S.A.M.; Birami, B.; Link, R.; Nadal-Sala, D.; Schuldt, B.; Yakir, D.; Ruehr, N.K. Anatomical adjustments of the tree hydraulic pathway decrease canopy conductance under long-term elevated CO2. Plant Physiol. 2022, kiac482, 1–20. [Google Scholar] [CrossRef]
- Buraczyk, W.; Tulik, M.; Konecka, A.; Szeligowski, H.; Czacharowski, M.; Bedkwski, M. Does leaf mass per area (LMA) discriminate natural pine populations of different origins? Eur. J. For. Res. 2022, 141, 1177–1187. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Sheng, B.L.; Zhao, H.L.; Ma, L.B.; Lin, J.; Liu, C.Y.; Chen, X.G. Leaf characteristics of Ginkgo biloba L. J. Plant Genet. Resour. 2004, 1, 65–68. [Google Scholar] [CrossRef]
- Hu, P.T.; Yan, X.C.; Luo, J.X.; Liu, F.R.; Du, Y.P. Phenotypic variation of Ginkgo leaves in Sichuan native land. J. Sichuan For. Sci. Technol. 2020, 41, 64–70. [Google Scholar]
- Kondratskaya, E.L.; Krishtal, O.A. Effects of Ginkgo biloba extract constituents on glycine-activated strychnine-sensitive receptors in hippocampal pyramidal neurons of the rat. Neurophysiology 2002, 2, 155–157. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Wu, Q.; Yao, X.; Cheng, Z.Q. Rapid and sensitive determination of major active ingredients and toxic components in Ginkgo biloba leaves extract (EGb 761) by a validated UPLC–MS-MS Method. J. Chromatogr. Sci. 2017, 4, 459–464. [Google Scholar] [CrossRef] [Green Version]
- González-Martínez, S.C.; Ersoz, E.; Brown, G.R.; Wheeler, N.C.; Neale, D.B. DNA sequence variation and selection of tag single-nucleotide polymorphisms at candidate genes for drought-stress response in Pinus taeda L. Genetics 2006, 172, 1915–1926. [Google Scholar] [CrossRef] [Green Version]
- González-Martínez, S.C.; Wheeler, N.C.; Ersoz, E.; Nelson, C.D.; Neale, D.B. Association genetics in Pinus taeda L. I. Wood property traits. Genetics 2007, 175, 399–409. [Google Scholar] [CrossRef] [Green Version]
- Thumma, B.R.; Nolan, M.F.; Evans, R.; Moran, G.F. Polymorphisms in Cinnamoyl CoA Reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 2005, 171, 1257–1265. [Google Scholar] [CrossRef] [Green Version]
- Ingvarsson, P.K.; Garcia, M.V.; Luquez, V.; Hall, D.; Jansson, S. Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 2008, 178, 2217–2226. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Mu, K.; Ni, Z.; Liu, X.; Li, Y.; Xu, L.A. Analysis of genetic diversity of ancient Ginkgo populations using SSR markers. Ind. Crops Prod. 2020, 145, 111942. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Burbano, H.A.; Hodges, E.; Green, R.E.; Briggs, A.W.; Krause, J.; Meyer, M.; Good, J.M.; Maricic, T.M.; Johnson, P.L.F.; Xuan, Z.Y.; et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science 2010, 328, 723–725. [Google Scholar] [CrossRef] [Green Version]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.A.; Bhangoo, J.; Fernández-Fernández, F.; Moore, P.; Swanson, J.D.; Viola, R.; Velasco, R.; Bassil, N.; Weber, C.A.; Sargent, D.J. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genom. 2013, 14, 2. [Google Scholar] [CrossRef]
- Huang, Y.F.; Poland, J.A.; Wight, C.P.; Jackson, E.W.; Tinker, N.A. Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PLoS ONE 2014, 9, e102448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto, J.C.; Ortiz, J.F.; Perlaza-Jiménez, L.; Vásquez, A.X.; Augusto, L.; Lopez-Lavalle, B.; Mathew, B.; Léon, J.; Bernal, A.J.; Ballvora, A.; et al. A genetic map of cassava (Manihot esculenta Crantz) with integrated physical mapping of immunity-related genes. BMC Genom. 2015, 16, 190. [Google Scholar] [CrossRef] [Green Version]
- Guan, R.; Zhao, Y.P.; Zhang, H.; Fan, G.Y.; Liu, X.; Zhou, W.B.; Shi, C.C.; Wang, J.H.; Liu, W.Q.; Liang, X.M.; et al. Draft genome of the living fossil Ginkgo biloba. Gigascience 2016, 5, 49. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Narasimhan, V.; Danecek, P.; Scally, A.; Xue, Y.L.; Tyler-Smith, C.; Durbin, R. BCFtools/RoH: A hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 2016, 32, 1749–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wang, X.; Wang, G.; Cui, P.; Wu, S.G.; Ai, C.; Hu, N.; Li, A.L.; He, B.; Shao, X.J.; et al. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Lee, U.; Mortola, E.; Kim, E.; Long, M. Evolution and maintenance of phenotypic plasticity. Biosystems 2022, 222, 104791. [Google Scholar] [CrossRef]
- Guo, Y.; Lu, Y.; El-Kassaby, Y.A.; Feng, L.; Wang, G.B.; Wang, T.L. Predicting growth and habitat responses of Ginkgo biloba L. to climate change. Ann. For. Sci. 2019, 76, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Li, X.F.; Chen, X.L. Analysis on phenotypic characters and diversity of Ginkgo biloba leaves in Zhejiang Province. Zhejiang J. Tradit. Chin. Med. 2017, 52, 538–539. [Google Scholar]
- Zuo, L.H.; Zhang, J.; Dong, Y.; Wang, J.M.; Ren, T.C. Genetic diversity of Malus sieversu natural population from Xinjiang analyzed by leaf traits. Nor. Horticult. 2015, 39, 1–7. [Google Scholar]
- Li, M.W.; Zhu, Z.H.; Wang, A.D.; Yao, X.F.; Yang, Z.S.; Cheng, D.S.; Yin, S.X. Leaf shape variation and Hybrid purity identification of several watermelon populations. J. Anhui Agr. Sci. 2001, 2, 213–216. [Google Scholar]
- Hui, L.X. Genetic Diversity and Phytogeography of Liriodendron Chinese. Ph.D. Thesis, Nanjing Forestry University, Nanjing, China, 2010. [Google Scholar]
- Li, Y.Q.; Zou, D.T.; Shrestha, N.; Xu, X.T.; Wang, Q.G.; Jia, W.; Wang, Z.H. Spatiotemporal variation in leaf size and shape in response to climate. J. Plant Ecol. 2020, 13, 87–96. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Leigh, A.; Boyce, C.K.; Jones, C.S.; Niklas, K.J.; Royer, D.L.; Tsukaya, H. The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 2011, 38, 535–552. [Google Scholar] [CrossRef] [Green Version]
- Royer, D.L.; McElwain, J.C.; Adams, J.M.; Wilf, P. Sensitivity of leaf size and shape to climate within Acer rubrum and Quercus kelloggii. New Phytol. 2008, 179, 808–817. [Google Scholar] [CrossRef]
- Huang, W.J.; Li, Z.J.; Yang, Z.P.; Bai, G.Z. The structural traits of Populus euphratica heteromorphic leaves and their correlations. Acta Ecol. Sin. 2010, 30, 4636–4642. [Google Scholar]
- Zhao, Y.P.; Fan, G.Y.; Yin, P.P.; Sun, S.; Li, N.; Hong, X.N.; Hu, G.; Zhang, H.; Zhang, F.M.; Han, J.D.; et al. Resequencing 545 ginkgo genomes across the world reveals the evolutionary history of the living fossil. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Matesanz, S.; Gianoli, E.; Valladares, F. Global change and the evolution of phenotypic plasticity in plants. Ann. N. Y. Acad. Sci. 2010, 1206, 35–55. [Google Scholar] [CrossRef]
- Kuhn, B.M.; Geisler, M.; Bigler, L.; Ringli, C. Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 2011, 156, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Gao, C.Y.; Wang, M.K.; Fu, F.F.; EI-Kassaby, Y.A.; Wang, T.L.; Wang, G.B. Metabolome and transcriptome analyses reveal flavonoids biosynthesis differences in Ginkgo biloba associated with environmental conditions. Ind. Crops Prod. 2020, 158, 112963. [Google Scholar] [CrossRef]
- Brunetti, C.; Di Ferdinando, M.; Fini, A.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants and developmental regulators: Relative significance in plants and humans. Int. J. Mol. Sci. 2013, 14, 3540–3555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Q.G.; Du, R.; Gou, J.B.; Guo, L.J.; Shen, H.; Liu, H.L.; Nguyen, J.K.; Ming, R.; Yin, T.M.; Huang, S.W. The genomic architecture of the sex-determining region and sex-related metabolic variation in Ginkgo biloba. Plant J. 2020, 104, 1399–1409. [Google Scholar] [CrossRef]
- Shelton, A.L. Variable chemical defences in plants and their effects on herbivore behaviour. Evol. Ecol. Res. 2000, 2, 231–249. [Google Scholar]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of plant secondary metabolites to environmental factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.B.; Wang, L.; Pang, S.Y.; Jia, Z.C.; Wang, L.; Li, W.X.; Jin, B. UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind. Crops Prod. 2020, 151, 112483. [Google Scholar] [CrossRef]
- Liu, C.G.; Zhou, X.Q.; Chen, D.G.; Li, L.J.; Li, J.C.; Chen, T.D. Natural variation of leaf thickness and its association to yield traits in indica rice. J. Integr. Agr. 2014, 13, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.; He, S.; Sun, G.; Jia, Y.; Sun, Y.; Ma, W.; Dev, W.; Nazir, M.F.; Geng, X.; Wang, L.; et al. Integrating Genome-wide association and whole transcriptome analysis to reveal genetic control of leaf traits in Gossypium arboreum L. Genomics 2022, 114, 110331. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska, J.; Xu, J.; Seifertová, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef] [Green Version]
- Drost, D.R.; Puranik, S.; Novaes, E.; Novaes, C.R.D.B.; Dervinis, C.; Gailing, O.; Kirst, M. Genetical genomics of Populus leaf shape variation. BMC Plant Biol. 2015, 15, 166. [Google Scholar] [CrossRef] [PubMed]

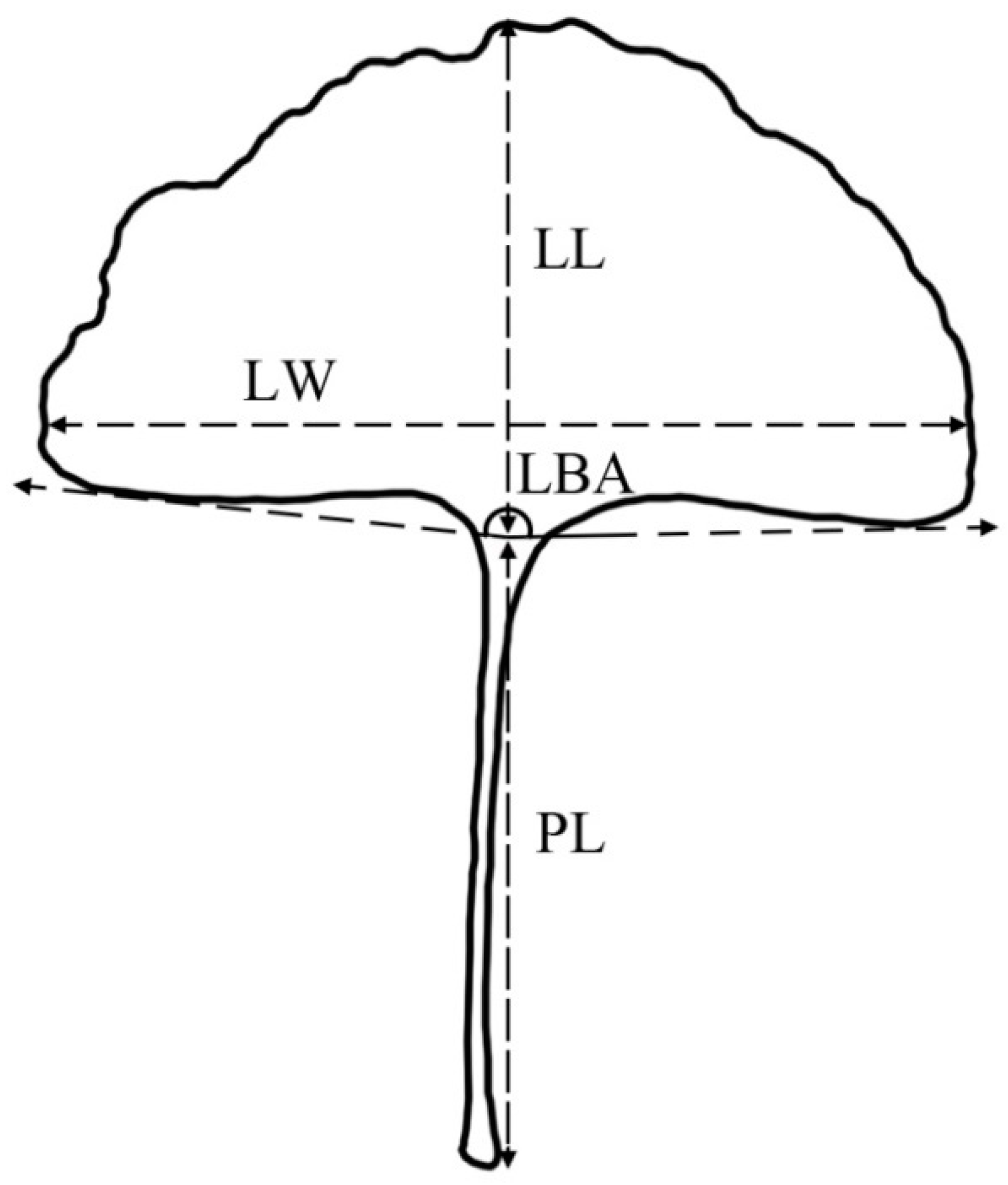
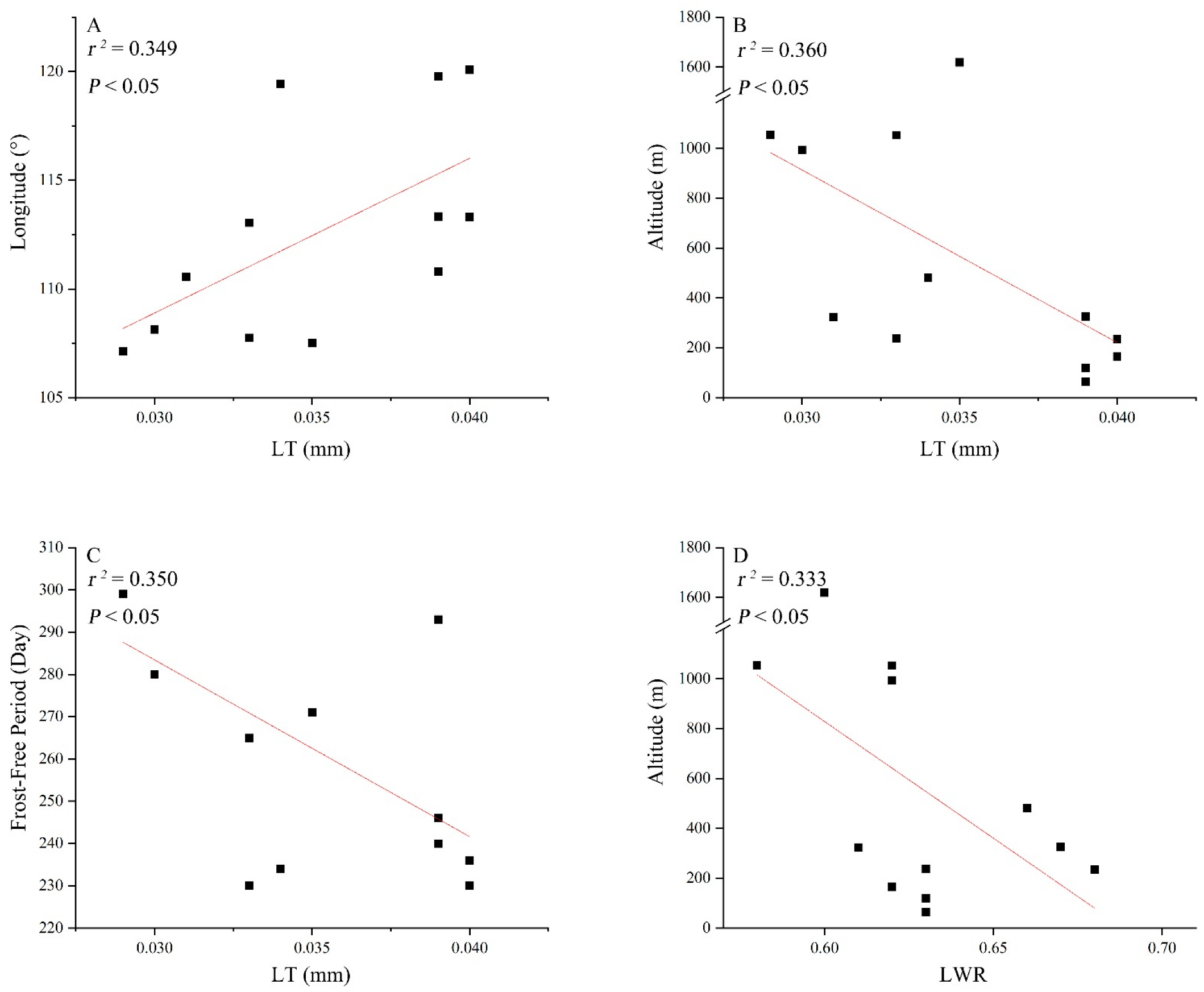
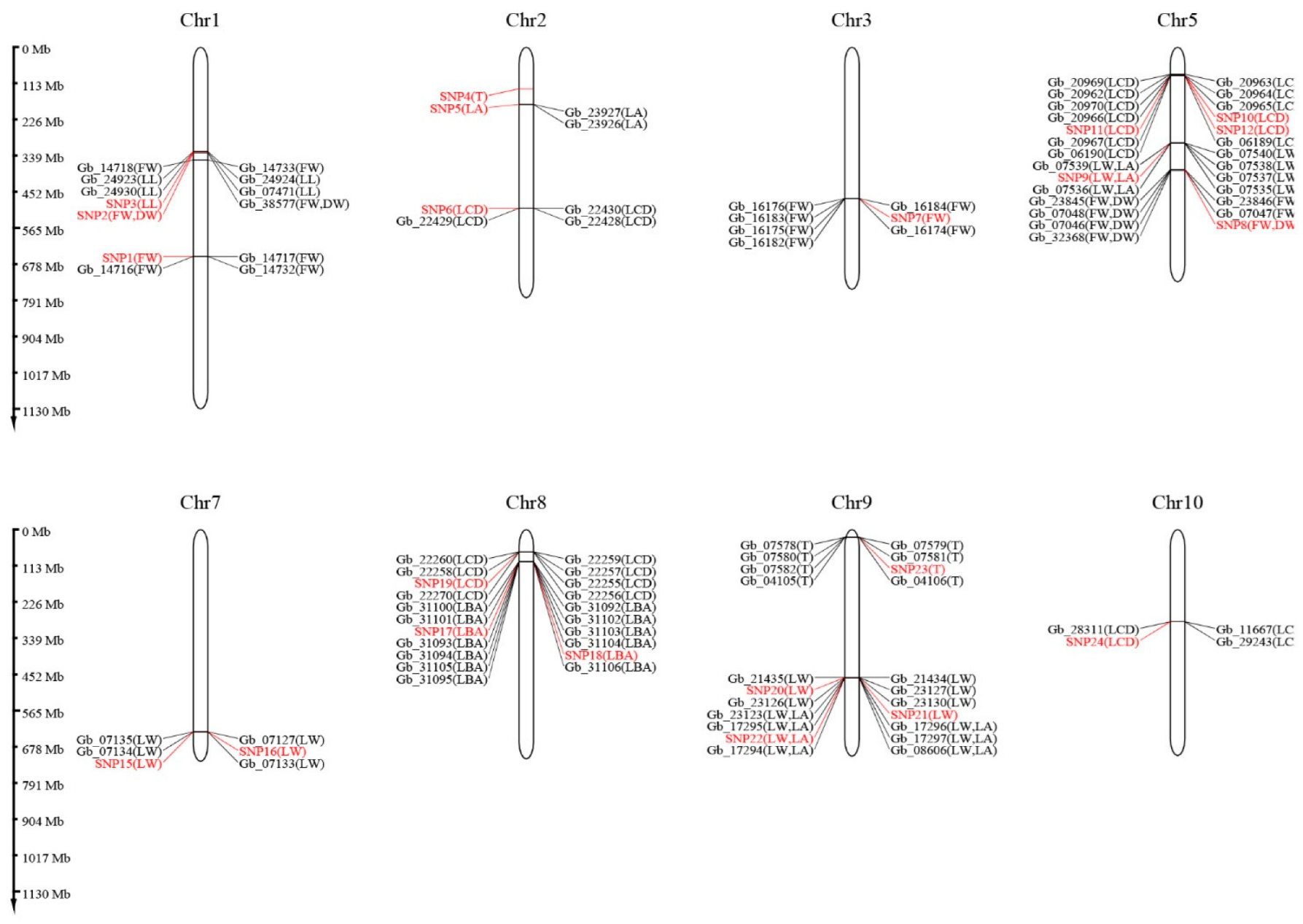

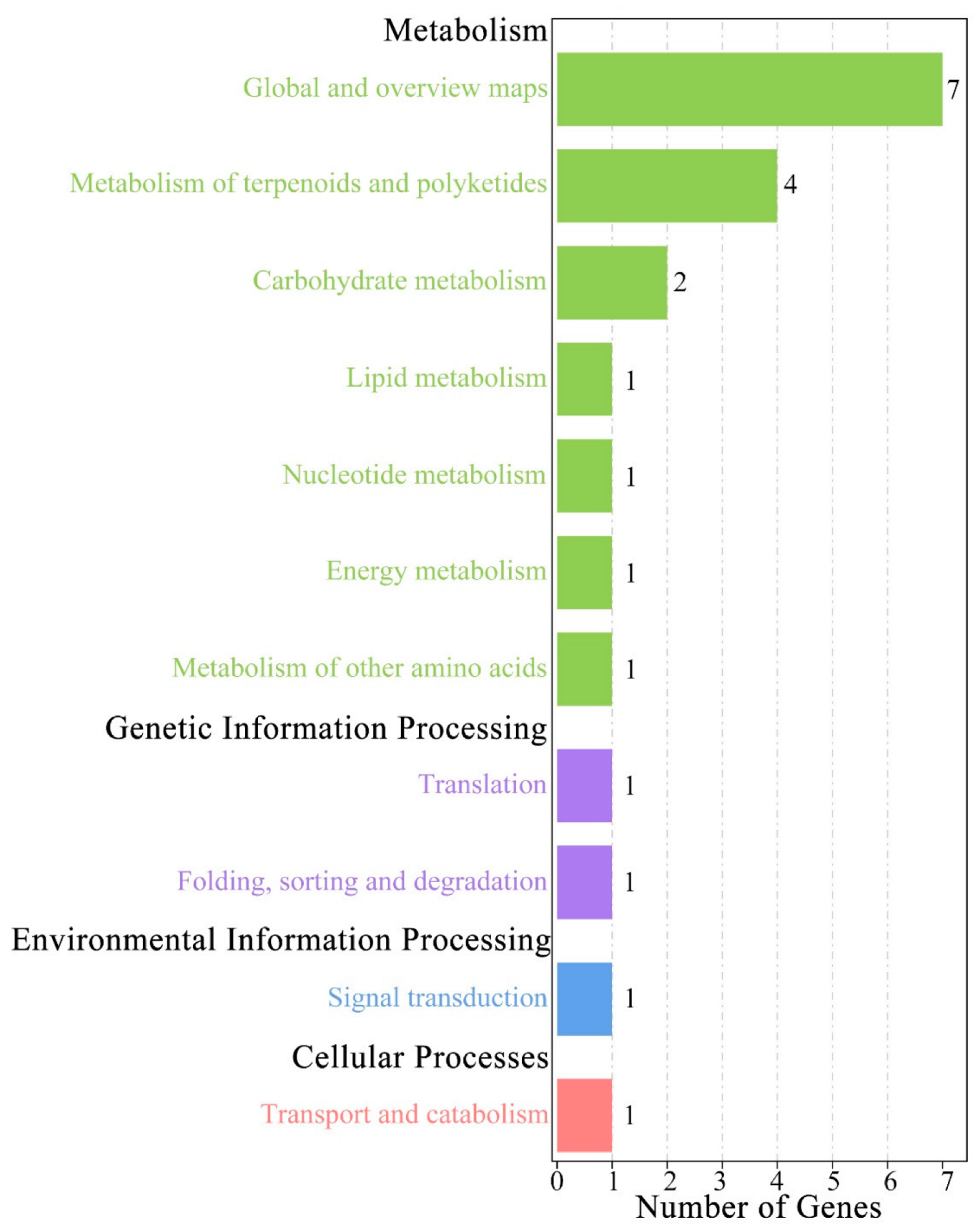
| Population Code | Location | Longitude (° E) | Latitude (° N) | Altitude (m) | Frost-Free Period (Day) | Annual Rainfall (mm) | N1 | N2 |
|---|---|---|---|---|---|---|---|---|
| PX | Panxian, Guizhou | 104.5 | 25.5 | 1619 | 271 | 1390 | 32 | 15 |
| DY | Duyun, Guizhou | 107.4 | 26.4 | 1054 | 299 | 1431 | 10 | 10 |
| FG | Fenggang, Guizhou | 107.8 | 27.8 | 1053 | 265 | 1200 | 14 | 14 |
| WC | Wuchuan, Guizhou | 108.1 | 28.6 | 994 | 280 | 1272 | 23 | 14 |
| LC | Lingchuan, Guangxi | 110.6 | 25.3 | 323 | 318 | 1926 | 28 | 14 |
| MC | Mochuan, Guangxi | 110.8 | 25.5 | 325 | 293 | 1842 | 29 | 0 |
| JS | Jingshan, Hubei | 113.1 | 31.3 | 238 | 230 | 1085 | 31 | 15 |
| SZ | Suizhou, Hubei | 113.3 | 31.4 | 235 | 230 | 968 | 30 | 15 |
| AL | Anlu, Hubei | 113.3 | 31.4 | 120 | 246 | 1100 | 33 | 0 |
| TM | Mt. Tianmu, Zhejiang | 119.4 | 30.3 | 481 | 234 | 956 | 34 | 14 |
| CX | Changxing, Zhejiang | 119.8 | 31.0′ | 64 | 240 | 1309 | 31 | 0 |
| ZJ | Zhuji, Zhejiang | 120.1 | 28.8 | 166 | 236 | 1374 | 26 | 15 |
| Total | 321 | 126 |
| FW **/g | DW **/g | LL **/cm | LW **/cm | LT **/mm | LA **/cm2 | PL **/cm | LBA **/° | LWR ** | |
|---|---|---|---|---|---|---|---|---|---|
| PX | 0.87 ± 0.18 cb/BA | 0.34 ± 0.09 b/A | 4.70 ± 0.52 cb/CB | 7.86 ± 0.81 dcb/CB | 0.35 ± 0.00 b/B | 23.68 ± 4.76 c/CB | 3.82 ± 0.64 dc/DCB | 162.92 ± 18.50 ba/CBA | 0.60 ± 0.03 ed/ED |
| DY | 0.47 ± 0.14 g/E | 0.16 ± 0.05 e/D | 3.90 ± 0.38 e/D | 6.70 ± 0.59 g/F | 0.29 ± 0.00 e/E | 17.42 ± 3.70 e/E | 3.99 ± 0.55 bc/BC | 148.74 ± 11.10 bc/BCD | 0.58 ± 0.03 e/E |
| FG | 0.67 ± 0.09 ef/CD | 0.21 ± 0.03 d/CD | 4.38 ± 0.38 cd/BC | 7.10 ± 0.37 efg/DEF | 0.33 ± 0.00 bcd/BCDE | 22.38 ± 1.92 cd/CD | 4.35 ± 0.80 ab/AB | 149.23 ± 15.50 bc/BCD | 0.62 ± 0.06 cd/CDE |
| WC | 0.57 ± 0.17 fg/DE | 0.20 ± 0.06 de/CD | 4.23 ± 0.66 de/CD | 6.85 ± 1.05 g/EF | 0.30 ± 0.00 cde/CDE | 18.53 ± 5.73 e/EF | 3.39 ± 0.70 de/D | 160.73 ± 19.08 ab/ABCD | 0.62 ± 0.03 cd/CDE |
| LC | 0.76 ± 0.21 cde/BC | 0.28 ± 0.08 c/B | 4.68 ± 0.64 bc/BC | 7.65 ± 1.02 cd/BCD | 0.31 ± 0.00 de/DE | 23.37 ± 5.76 cB/CD | 4.04 ± 0.58 bc/ABC | 166.11 ± 23.40 a/AB | 0.61 ± 0.03 cde/DE |
| MC | 0.99 ± 0.21 a/A | 0.37 ± 0.07 ab/A | 5.40 ± 0.75 a/A | 8.14 ± 0.67 bc/AB | 0.39 ± 0.00 a/A | 26.69 ± 4.49 b/B | 3.62 ± 0.74 cde/CD | 149.91 ± 28.63 bc/BCD | 0.67 ± 0.08 a/AB |
| JS | 0.76 ± 0.21 cde/BC | 0.27 ± 0.07 c/B | 4.69 ± 0.53 bc/BC | 7.43 ± 0.86 def/CDE | 0.33 ± 0.00 bc/BCD | 22.20 ± 4.94 cd/CDE | 3.84 ± 0.72 c/BCD | 161.12 ± 29.12 ab/ABCD | 0.63 ± 0.06 bc/BCD |
| SZ | 0.88 ± 0.21 bc/AB | 0.36 ± 0.08 ab/A | 5.58 ± 0.86 a/A | 8.21 ± 0.98 ab/AB | 0.40 ± 0.01 a/A | 26.59 ± 5.46 b/B | 3.38 ± 0.90 e/D | 143.04 ± 21.78 c/D | 0.68 ± 0.08 a/A |
| AL | 0.76 ± 0.15 cde/BC | 0.26 ± 0.05 c/BC | 4.78 ± 0.44 b/B | 7.58 ± 0.7 de/BCD | 0.39 ± 0.00 a/A | 23.06 ± 3.59 c/BCD | 3.69 ± 0.46 cde/CD | 162.24 ± 17.09 ab/ABC | 0.63 ± 0.05 bc/BCD |
| TM | 0.86 ± 0.19 bcd/AB | 0.34 ± 0.11 b/A | 4.62 ± 0.48 bc/BC | 7.00 ± 0.50 fg/DEF | 0.34 ± 0.00 b/BC | 19.50 ± 2.99 de/DEF | 3.97 ± 0.61 bc/BC | 145.82 ± 22.05 c/CD | 0.66 ± 0.04 ab/ABC |
| CX | 0.75 ± 0.20 de/BC | 0.25 ± 0.07 c/BC | 4.73 ± 0.61 bc/BC | 7.57 ± 0.96 de/BCD | 0.39 ± 0.00 a/A | 21.97 ± 4.52 cd/CDE | 3.82 ± 0.64 cd/BCD | 150.57 ± 17.5 bc/BCD | 0.63 ± 0.04 cd/BCD |
| ZJ | 0.93 ± 0.13 ab/A | 0.39 ± 0.07 a/A | 5.37 ± 0.54 a/A | 8.66 ± 0.87 a/A | 0.40 ± 0.00 a/A | 31.03 ± 5.36 a/A | 4.56 ± 0.66 a/A | 172.93 ± 17.86 a/A | 0.62 ± 0.03 cd/CDE |
| Mean | 0.77 | 0.29 | 4.76 | 7.56 | 0.35 | 23.03 | 3.87 | 156.11 | 0.63 |
| CV% | 18.60 | 24.84 | 9.88 | 7.48 | 10.90 | 15.71 | 8.56 | 5.73 | 4.27 |
| FW | DW | LL | LW | LT | LA | PL | LBA | LWR | Mean | |
|---|---|---|---|---|---|---|---|---|---|---|
| PX | 20.15 | 27.39 | 11.15 | 10.36 | 5.83 | 20.10 | 16.65 | 11.35 | 4.45 | 14.16 |
| DY | 29.54 | 30.88 | 9.76 | 8.76 | 11.58 | 21.25 | 13.89 | 7.47 | 5.59 | 15.41 |
| FG | 13.08 | 12.90 | 8.62 | 5.26 | 7.75 | 8.57 | 18.40 | 10.39 | 9.91 | 10.54 |
| WC | 30.39 | 29.74 | 15.68 | 15.30 | 12.16 | 30.94 | 20.70 | 11.87 | 4.85 | 19.07 |
| LC | 27.01 | 28.79 | 13.65 | 13.37 | 9.06 | 24.65 | 14.34 | 14.09 | 4.56 | 16.61 |
| MC | 21.35 | 18.81 | 13.96 | 8.26 | 9.00 | 16.83 | 20.43 | 19.10 | 12.23 | 15.55 |
| JS | 27.61 | 26.58 | 11.24 | 11.57 | 9.18 | 22.24 | 18.84 | 18.07 | 9.17 | 17.17 |
| SZ | 24.49 | 22.21 | 15.35 | 11.93 | 13.40 | 20.55 | 26.67 | 15.23 | 11.13 | 17.88 |
| AL | 19.58 | 20.30 | 9.16 | 9.24 | 12.32 | 15.56 | 12.48 | 10.53 | 7.99 | 13.02 |
| TM | 21.71 | 30.59 | 10.42 | 7.18 | 6.38 | 15.34 | 15.44 | 15.12 | 6.70 | 14.32 |
| CX | 27.15 | 27.03 | 12.85 | 12.65 | 9.92 | 20.58 | 16.87 | 11.62 | 5.80 | 16.05 |
| ZJ | 13.49 | 17.89 | 9.99 | 9.99 | 9.30 | 17.27 | 14.51 | 10.33 | 4.07 | 11.87 |
| Mean | 22.96 | 24.43 | 11.82 | 10.32 | 9.66 | 19.49 | 17.43 | 12.93 | 7.20 |
| Clean Data | Mapping Rate% | Q20 | Q30 | SNP Calling Rate% | |
|---|---|---|---|---|---|
| PX | 2.68 ± 0.58 | 99.78 ± 0.01 | 95.49 ± 0.52 | 88.88 ± 1.15 | 99.37 ± 0.01 |
| DY | 2.58 ± 0.48 | 99.67 ± 0.13 | 95.23 ± 0.73 | 88.27 ± 1.64 | 99.36 ± 0.01 |
| FG | 2.49 ± 0.51 | 99.62 ± 0.06 | 94.97 ± 0.43 | 87.87 ± 0.92 | 99.28 ± 0.01 |
| WC | 2.81 ± 0.48 | 99.71 ± 0.05 | 95.51 ± 0.65 | 88.89 ± 1.45 | 99.26 ± 0.01 |
| LC | 2.73 ± 0.76 | 99.63 ± 0.16 | 95.73 ± 0.42 | 89.39 ± 1 | 99.23 ± 0.01 |
| JS | 2.42 ± 0.41 | 99.67 ± 0.07 | 95.75 ± 0.35 | 89.39 ± 0.82 | 99.28 ± 0.01 |
| SZ | 2.79 ± 0.47 | 99.74 ± 0.04 | 96.02 ± 0.25 | 90 ± 0.65 | 99.33 ± 0.01 |
| TM | 2.8 ± 0.38 | 99.73 ± 0.07 | 95.67 ± 0.56 | 89.19 ± 1.28 | 99.26 ± 0.01 |
| ZJ | 2.28 ± 0.31 | 99.73 ± 0.07 | 95.13 ± 0.54 | 88.14 ± 1.17 | 99.38 ± 0.00 |
| Mean | 2.62 | 99.70 | 95.51 | 88.92 | 99.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Shen, X.; Li, Y. Variation Pattern and Genome-Wide Association Study of Leaf Phenotypic Traits among Ancient Ginkgo biloba L. Populations. Forests 2022, 13, 1764. https://doi.org/10.3390/f13111764
Zhou Q, Shen X, Li Y. Variation Pattern and Genome-Wide Association Study of Leaf Phenotypic Traits among Ancient Ginkgo biloba L. Populations. Forests. 2022; 13(11):1764. https://doi.org/10.3390/f13111764
Chicago/Turabian StyleZhou, Qi, Xin Shen, and Yingang Li. 2022. "Variation Pattern and Genome-Wide Association Study of Leaf Phenotypic Traits among Ancient Ginkgo biloba L. Populations" Forests 13, no. 11: 1764. https://doi.org/10.3390/f13111764
APA StyleZhou, Q., Shen, X., & Li, Y. (2022). Variation Pattern and Genome-Wide Association Study of Leaf Phenotypic Traits among Ancient Ginkgo biloba L. Populations. Forests, 13(11), 1764. https://doi.org/10.3390/f13111764






