Changes in Chemical Properties and Fungal Communities of Mineral Soil after Clear-Cutting and Reforestation of Scots Pine (Pinus sylvestris L.) Sites
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Soil Sampling and Chemical Analyses
2.3. Fungal Sampling
2.4. Fungal DNA Isolation, Amplification, and Sequencing
2.5. Bioinformatics
2.6. Statistical Analyses
3. Results
3.1. Chemical Properties of Mineral Topsoil
3.2. Fungal Community
3.3. Relations between Chemical Parameters and Fungal Community in the Mineral Topsoil
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gustafsson, L.; Baker, S.C.; Bauhus, J.; Beese, W.J.; Brodie, A.; Kouki, J.; Lindenmayer, D.B.; Lohmus, A.; Pastur, G.M.; Messier, C.; et al. Retention forestry to maintain multifunctional forests: A world perspective. Bioscience 2012, 62, 633–645. [Google Scholar] [CrossRef] [Green Version]
- Kuuluvainen, T. Disturbance dynamics in boreal forests: Defining the ecological basis of restoration and management of biodiversity. Silva Fenn. 2002, 36, 5–11. [Google Scholar] [CrossRef]
- Chazdon, R.L. Beyond deforestation: Restoring forests and ecosystem services on degraded lands. Science 2008, 320, 1458–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuuluvainen, T. Forest Management and Biodiversity Conservation Based on Natural Ecosystem Dynamics in Northern Europe: The Complexity Challenge. Ambio 2009, 38, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Bouget, C.; Nusillard, B.; Pineau, X.; Ricou, C. Effect of deadwood position on saproxylic beetles in temperate forests and conservation interest of oak snags. Insect Conserv. Divers. 2012, 5, 264–278. [Google Scholar] [CrossRef]
- Koivula, M.J. Useful model organisms, indicators, or both? Ground beetles (Coleoptera, Carabidae) reflecting environmental conditions. Zookeys 2011, 100, 287–317. [Google Scholar] [CrossRef] [Green Version]
- Sklodowski, J.J. Manual soil preparation and piles of branches can support ground beetles (Coleoptera, carabidae) better than four different mechanical soil treatments in a clear-cut area of a closed-canopy pine forest in northern Poland. Scand. J. For. Res. 2016, 32, 123–133. [Google Scholar] [CrossRef]
- Junninen, K.; Penttila, R.; Martikainen, P. Fallen retention aspen trees on clear-cuts can be important habitats for red-listed polypores: A case study in Finland. Biodivers. Conserv. 2007, 16, 475–490. [Google Scholar] [CrossRef]
- Paillet, Y.; Berges, L.; Hjalten, J.; Odor, P.; Avon, C.; Romermann, B.M.; Bijlsma, R.J.; De Bruyn, L.; Fuhr, M.; Grandin, U.; et al. Biodiversity Differences between Managed and Unmanaged Forests: Meta-Analysis of Species Richness in Europe. Conserv. Biol. 2009, 24, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Burivalova, Z.; Koh, L.P.; Hellweg, S. Impact of Forest Management on Species Richness: Global Meta-Analysis and Economic Trade-Offs. Sci. Rep. 2016, 6, 23954. [Google Scholar] [CrossRef]
- Chandler, J.R.; Haeussler, S.; Hamilton, E.H.; Feller, M.; Bradfield, G.; Simard, S.W. Twenty years of ecosystem response after clearcutting and slashburning in conifer forests of central British Columbia, Canada. PLoS ONE 2017, 12, e0172667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 2nd ed.; Academic Press: London, UK, 1997; p. 605. [Google Scholar]
- Klaubauf, S.; Inselsbacher, E.; Zechmeister-Boltenstern, S.; Wanek, W.; Gottsberger, R.; Strauss, J.; Gorfer, M. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fungal Divers. 2010, 44, 65–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menkis, A.; Uotila, A.; Arhipova, N.; Vasaitis, R. Effects of stump and slash removal on growth and mycorrhization of Picea abies seedlings outplanted on a forest clear-cut. Mycorrhiza 2010, 20, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.P.; Geisen, S. Trophic Regulations of the Soil Microbiome. Trends Microbiol. 2019, 27, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Behnke-Borowczyk, J.; Kowalkowski, W.; Kartawik, N.; Baranowska, M.; Barzdajn, W. Soil fungal communities in nurseries producing Abies alba. Balt. For. 2020, 26, 5–12. [Google Scholar] [CrossRef]
- Clarke, N.; Podenphant, L.K.; Kjonaas, O.J.; Barcena, T.G.; Vesterdal, L.; Stupak, I.; Finer, L.; Jacobson, S.; Armolaitis, K.; Lazdina, D.; et al. Effects of intensive biomass harvesting on forest soils in the Nordic countries and the UK: A meta-analysis. For. Ecol. Manag. 2021, 482, 118877. [Google Scholar] [CrossRef]
- Simard, S.W.; Austin, M.E. The Role of Mycorrhizas in Forest Soil Stability with Climate Change. In Climate Change and Variability; Simard, S.W., Austin, M.E., Eds.; IntechOpen: Rijeka, Croatia, 2010; Chapter 15; pp. 275–303. [Google Scholar]
- Kucerava, B.; Dobrovolny, L.; Remes, J. Responses of Abies alba seedlings to different site conditions in Picea abies plantations. Dendrobiology 2013, 69, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Kobala, M.; Grčman, H.; Zupan, M.; Levanič, T.; Simončič, P.; Kadunc, A.; Hladnik, D. Influence of soil properties on silver fir (Abies alba Mill.) growth in the Dinaric Mountains. For. Ecol. Manag. 2015, 337, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Kormanek, M.; Banach, J.; Lenczuk, D. Influence of soil compaction on the growth of silver fir (Abies alba Mill.) under a forest canopy. Ecol. Quest. 2015, 22, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Delgado, E.F.; Valdez, A.T.; Covarrubias, S.A.; Tosi, S.; Nicola, L. Soil Fungal Diversity of the Aguarongo Andean Forest (Ecuador). Biology 2021, 10, 1289. [Google Scholar] [CrossRef] [PubMed]
- Cairney, J.W.G.; Meharg, A.A. Interactions between ectomycorrhizal fungi and soil saprotrophs: Implications for decomposition of organic matter in soils and degradation of organic pollutants in the rhizosphere. Can. J. Bot. 2002, 80, 803–809. [Google Scholar] [CrossRef]
- Hobbie, E.A.; Macko, S.A.; Shugart, H.H. Insights into nitrogen and carbon dynamics of ectomycorrhizal and saprotrophic fungi from isotopic evidence. Oecologia 1999, 118, 353–360. [Google Scholar] [CrossRef]
- Jones, M.D.; Durall, D.M.; Cairney, J.W.G. Ectomycorrhizal fungal communities in young forest stands regenerating after clearcut logging. N. Phytol. 2003, 157, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teste, F.P.; Karst, J.; Jones, M.D.; Simard, S.W.; Durall, D.M. Methods to control ectomycorrhizal colonization: Effectiveness of chemical and physical barriers. Mycorrhiza 2006, 17, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Luoma, D.L.; Eberhart, J.L.; Molina, R.; Amaranthus, M.P. Response of ectomycorrhizal fungus sporocarp production to varying levels and patterns of green-tree retention. For. Ecol. Manag. 2004, 202, 337–354. [Google Scholar] [CrossRef]
- Nehls, U. Mastering ectomycorrhizal symbiosis: The impact of carbohydrates. J. Exp. Bot. 2008, 59, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Byrd, K.B.; Parker, V.T.; Vogler, D.R.; Cullings, K.W. The influence of clear-cutting on ectomycorrhizal fungus diversity in a lodgepole pine (Pinus contorta) stand, Yellowstone National Park, Wyoming, and Gallatin National Forest, Montana. Can. J. Bot. 2000, 78, 149–156. [Google Scholar]
- Lithuanian State Forest Survey Service. In Lithuanian Statistical Yearbook of Forestry; Lithuanian State Forest Survey Service: Kaunas, Lithuania, 2020; p. 25.
- Rudawska, M.; Leski, T. Ectomycorrhizal Fungal Assemblages of Nursery-Grown Scots Pine are Influenced by Age of the Seedlings. Forests 2021, 12, 134. [Google Scholar] [CrossRef]
- Wubet, T.; Christ, S.; Schöning, I.; Boch, S.; Gawlich, M.; Schnabel, B.; Fischer, M.; Buscot, F. Differences in soil fungal communities between European Beech (Fagus sylvatica L.) dominated forests are related to soil and understory vegetation. PLoS ONE 2012, 7, e47500. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Kemler, M.; Begerow, D. Assessment of yeast diversity in soils under different management regimes. Fungal Ecol. 2012, 5, 24–35. [Google Scholar] [CrossRef]
- Goldmann, K.; Schöning, I.; Buscot, F.; Wubet, T. Forest management type influences diversity and community composition of soil fungi across temperate forest ecosystems. Front. Microbiol. 2015, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Buee, M.; Reich, M.; Murat, C.; Morin, E.; Nilsson, R.H.; Uroz, S.; Martin, F. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009, 184, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Kwasna, H.; Walkowiak, L.; Lakomy, P.; Behnke-Borowczyk, J.; Gornowicz, R.; Mikicinski, A.; Gałązka, S.; Szewczyk, W. Effect of silvicultural techniques on the diversity of microorganisms in forest soil and their participation in biological control of Armillaria and Heterobasidion. J. Plant Prot. Res. 2015, 55, 241–253. [Google Scholar] [CrossRef]
- Malecka, M.; Kwasna, H.; Szewczyk, W. Fungal communities in barren forest soil after amendment with different wood substrates and their possible effects on trees pathogens, insects and nematodes. J. Plant Prot. Res. 2015, 55, 301–311. [Google Scholar] [CrossRef]
- Aučina, A.; Rudawska, M.; Leski, T.; Skridaila, A.; Riepšas, E.; Iwanski, M. Growth and mycorrhizal community structure of Pinus sylvestris seedlings following the addition of forest litter. Appl. Environ. Microbiol. 2007, 73, 4867–4873. [Google Scholar] [CrossRef] [Green Version]
- Aučina, A.; Rudawska, M.; Leski, T.; Skridaila, A.; Pašakinskiene, I.; Riepšas, E. Forest litter as the mulch improving growth and ectomycorrhizal diversity of bare-root Scots pine (Pinus sylvestris) seedlings. Iforest 2015, 8, 394–400. Available online: http://www.sisef.it/iforest/contents/?id=ifor1083-008 (accessed on 25 August 2022).
- Horton, T.R.; Bruns, T.D. The molecular revolution in ectomycorrhizal ecology: Peeking into the black-box. Mol. Ecol. 2001, 10, 1855–1871. [Google Scholar] [CrossRef] [Green Version]
- Menkis, A.; Vasaitis, R. Fungi in roots of nursery grown Pinus sylvestris: Ectomycorrhizal colonisation, genetic diversity and spatial distribution. Microb. Ecol. 2011, 61, 52–63. [Google Scholar] [CrossRef]
- Banchi, E.; Ametrano, C.G.; Tordoni, E.; Stankovi, D.; Ongaro, S.; Tetiach, M.; Pallavicini, A.; Muggia, L.; Verardo, P.; Tassan, F.; et al. Environmental DNA assessment of airborne plant and fungal seasonal diversity. Sci. Total Environ. 2020, 738, 140249. [Google Scholar] [CrossRef]
- Marčiulynas, A.; Marčiulyniene, D.; Lynikiene, J.; Gedminas, A.; Vaičiukyne, M.; Menkis, A. Fungi and oomycetes in the irrigation water of forest nurseries. Forests 2020, 11, 459. [Google Scholar] [CrossRef] [Green Version]
- Marčiulyniene, D.; Marčiulynas, A.; Lynikiene, J.; Vaičiukyne, M.; Gedminas, A.; Menkis, A. DNA-Metabarcoding of Belowground Fungal Communities in Bare-Root Forest Nurseries: Focus on Different Tree Species. Microorganisms 2021, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Karazija, S. Lietuvos Miškų Tipai. (Forest tipes of Lithuania); Mokslas: Vilnius, Lithuania, 1988; p. 210. (In Lithuanian) [Google Scholar]
- Vaičys, M.; Karazija, S.; Kuliešis, A.; Rutkauskas, A. Miškų Augavietės (Forest Sites); Lutute: Kaunas, Lithuania, 2006; p. 95. (In Lithuanian) [Google Scholar]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soil Resources Reports No. 106; FAO: Rome, Italy, 2015; p. 192. [Google Scholar]
- Sampling and Analysis of Soil. 2010. Available online: https://www.icp-forests.org/pdf/manual/2010/Manual_2010_soil.pdf (accessed on 25 August 2022).
- Rosling, A.; Landeweert, R.; Lindah, B.D.; Larsson, K.H.; Kuyper, T.W.; Taylor, A.F.S.; Finlay, R.D. Vertical distribution of ectomycorrhizal fungal taxa in a podzol soil profile. New Phytol. 2003, 159, 775–783. [Google Scholar]
- Ihrmark, K.; Bodeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandstrom-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region-evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Magurran, A.E. Ecological Diversity and Its Measurement, 1st ed.; Princeton University Press: Princeton, NJ, USA, 1988; p. 192. [Google Scholar]
- Nguyen, D.; Boberg, J.; Ihrmark, K.; Stenstrom, E.; Stenlid, J. Do foliar fungal communities of Norway spruce shift along a tree species diversity gradient in mature European forests? Fungal Ecol. 2016, 23, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Jussy, J.H.; Ranger, J.; Bienaimé, S.; Dambrine, E. Effects of a clear-cut on the in situ nitrogen mineralisation and the nitrogen cycle in a 67-year old Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco) plantation. Ann. For. Sci. 2004, 61, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Gundersen, P.; Schmidt, I.K.; Raulund-Rasmussen, K. Leaching of nitrate from temperate forests—Effects of air pollution and forest management. Environ. Rev. 2006, 14, 1–57. [Google Scholar] [CrossRef]
- Hedwall, P.O.; Grip, H.; Linder, S.; Lövdahl, L.; Nilsson, U.; Bergh, J. Effects of clear-cutting and slash removal on soil water chemistry and forest floor vegetation in a nutrient optimised Norway spruce stand. Silva Fenn. 2013, 47, 16. [Google Scholar] [CrossRef] [Green Version]
- Palviainen, M. Logging residues and ground vegetation in nutrient dynamics of a clear-cut boreal forest. Diss. For. 2005, 12, 1–38. [Google Scholar] [CrossRef] [Green Version]
- Wall, A. Effect of removal of logging residue on nutrient leaching and nutrient pools in the soil after clearcutting in a Norway spruce stand. For. Ecol. Manag. 2008, 256, 1372–1383. [Google Scholar] [CrossRef]
- Armolaitis, K.; Varnagirytė-Kabašinskienė, I.; Stupak, I.; Mikšys, V.; Kukkola, M.; Wójcik, J. Carbon and nutrients of Scots pine stands on sandy soils in Lithuania in relation to bioenergy sustainability. Biomass Bioenergy 2013, 54, 250–259. [Google Scholar] [CrossRef]
- Smolander, A.; Kitunen, V.; Mälkönen, E. Dissolved soil organic nitrogen and carbon in a Norway spruce stand and adjacent clear-cut. Biol. Fertil. Soils 2001, 33, 190–196. [Google Scholar] [CrossRef]
- Palviainen, M.; Finér, L.; Laurén, A.; Mattsson, T.; Högbom, L. A method to estimate the impact of clear-cutting on nutrient concentrations in boreal headwater streams. Ambio 2015, 44, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Stuiver, B.; Wardle, D.; Gundale, M.; Nilsson, M. Seedling responses to changes in canopy and soil properties during stand development following clear-cutting. For. Ecol. Manag. 2016, 378, 31–43. [Google Scholar] [CrossRef]
- Rappe George, M.O.; Hansson, L.J.; Ring, E.; Jansson, P.E.; Gärdenäs, A.I. Nitrogen leaching following clear-cutting and soil scarification at a Scots pine site—A modelling study of a fertilization experiment. For. Ecol. Manag. 2017, 385, 281–294. [Google Scholar] [CrossRef]
- Česonienė, L.; Daubaras, R.; Tamutis, V.; Kaškonienė, V.; Kaškonas, P.; Stakėnas, V.; Zych, M. Effect of clear-cutting on the understory vegetation, soil and diversity of litter beetles in scots pine-dominated forest. J. Sustain. For. 2019, 38, 791–808. [Google Scholar] [CrossRef]
- Six, J.; Conant, R.T.; Paul, E.A. Stabilization mechanisms of soil organic matter: Implications for C—Saturation of soil. Plant Soil 2002, 241, 155–176. [Google Scholar] [CrossRef]
- Lal, R. Sequestering carbon in soils of agro-ecosystems. Food Policy 2011, 36, 33–39. [Google Scholar] [CrossRef]
- Qualls, R.G.; Haines, B.L.; Swank, W.T.; Tyler, S.W. Soluble organic and inorganic nutrient fluxes in clearcut and mature deciduous forests. Soil Sci. Soc. Am. J. 2000, 64, 1068–1077. [Google Scholar] [CrossRef] [Green Version]
- Piirainen, S.; Finer, L.; Mannerkoski, H.; Starr, M. Effects of forest clear-cutting on the carbon and nitrogen fluxes through podzolic soil horizons. Plant Soil 2002, 239, 301–311. [Google Scholar] [CrossRef]
- Schelker, J.; Eklöf, K.; Bishop, K.; Laudon, H. Effects of forestry operations on dissolved organic carbon concentrations and export in boreal first-order streams. J. Geophys. Res. 2012, 117, G01011. [Google Scholar] [CrossRef]
- Schelker, J.; Öhman, K.; Löfgren, S.; Laudon, H. Scaling of increased dissolved organic carbon inputs by forest clear-cutting—What arrives downstream? J. Hydrol. 2014, 508, 299–306. [Google Scholar] [CrossRef]
- Nieminen, M.; Sarkkola, S.; Laurén, A. Impacts of forest harvesting on nutrient, sediment and dissolved organic carbon exports from drained peatlands: A literature review, synthesis and suggestions for the future. For. Ecol. Manag. 2017, 392, 13–20. [Google Scholar] [CrossRef]
- Cools, N.; Vesterdal, L.; De Vos, B.; Vanguelova, E.; Hansen, K. Tree species is the major factor explaining C:N ratios in European forest soils. For. Ecol. Manag. 2014, 311, 3–16. [Google Scholar] [CrossRef]
- Siebers, N.; Kruse, J. Short-term impacts of forest clear-cut on soil structure and consequences for organic matter composition and nutrient speciation: A case study. PLoS ONE 2019, 14, e0220476. [Google Scholar] [CrossRef] [Green Version]
- Sterkenburg, E.; Clemmensen, K.E.; Lindahl, B.D.; Dahlberg, A. The significance of retention trees for survival of ectomycorrhizal fungi in clear−cut Scots pine forests. J. Appl. Ecol. 2019, 56, 1367–1378. [Google Scholar] [CrossRef]
- Orwin, K.H.; Kirschbaum, M.U.F.; St John, M.G.; Dickie, I.A. Organic nutrient uptake by mycorrhizal fungi enhances ecosystem carbon storage: A model-based assessment: Organic nutrient uptake enhances soil C. Ecol. Lett. 2011, 14, 493–502. [Google Scholar] [CrossRef]
- Averill, C.; Hawkes, C.V. Ectomycorrhizal fungi slow soil carbon cycling. Ecol. Lett. 2016, 19, 937–947. [Google Scholar] [CrossRef]
- Rosling, A.; Timling, I.; Taylor, D.L. Archaeorhizomycetes: Patterns of distribution and abundance in soil. In Genomics of Soil-and Plant-Associated Fungi; Horwitz, B.A., Mukherjee, P.K., Mukherjee, M., Kubicek, C.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 3, pp. 333–349. [Google Scholar] [CrossRef] [Green Version]
- Menkis, A.; Urbina, H.; James, T.Y.; Rosling, A. Archaeorhizomyces borealis sp. nov. and a sequence-based classification of related soil fungal species. Fungal Biol. 2014, 118, 943–955. [Google Scholar] [CrossRef]
- Choma, M.; Bárta, J.; Šantrucková, H.; Urich, T. Low abundance of Archaeorhizomycetes among fungi in soil metatranscriptomes. Sci. Rep. 2016, 6, 38455. [Google Scholar] [CrossRef]
- Lee, E.H.; Eom, A.H. Ectomycorrhizal fungal communities of red pine (Pinus densiflora) seedlings in disturbed sites and undisturbed old forest sites. Mycobiology 2013, 41, 77–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwasna, H.; Behnke-Borowczyk, J.; Gornowicz, R.; Lakomy, P. Effects of preparation of clear-cut forest sites on the soil mycobiota with consequences for Scots pine growth and health. For. Path. 2019, 49, 1–12. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above and below ground views. Can. J. Bot. 1996, 74, 1572–1583. [Google Scholar] [CrossRef]
- Dahlberg, A.; Jonsson, L.; Nylund, J.E. Species diversity and distribution of biomass above and below ground among ectomycorrhizal fungi in an old-growth Norway spruce forest in South Sweden. Can. J. Bot. 1997, 75, 1323–1335. [Google Scholar] [CrossRef]
- Teste, F.P.; Lieffers, V.J.; Strelkov, S.E. Ectomycorrhizal community responses to intensive forest management: Thinning alters impacts of fertilization. Plant Soil 2012, 360, 333–347. [Google Scholar] [CrossRef]
- Hartmann, M.; Howes, C.G.; VanInsberghe, D.; Yu, H.; Bachar, D.; Christen, R.; Nilsson, R.H.; Hallam, S.J.; Mohn, W.W. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J. 2012, 6, 2199–2218. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, B.J.; Jones, M.D.; Kranabetter, J.M. Ectomycorrhizae and tree seedling nitrogen nutrition in forest restoration. New For. 2015, 46, 747–771. [Google Scholar] [CrossRef]
- Policelli, N.; Horton, T.R.; Hudon, A.T.; Patterson, T.R.; Bhatnagar, J.M. Back to Roots: The Role of Ectomycorrhizal Fungi in Boreal and Temperate Forest Restoration. Front. For. Glob. Chang. 2020, 3, 97. [Google Scholar] [CrossRef]
- Futai, K.; Taniguchi, T.; Kataoka, R. Ectomycorrhizae and their importance in forest ecosystems. In Mycorrhizae: Sustainable Agriculture and Forestry; Siddiqui, Z., Akhtar, M.S., Futai, K., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 241–285. [Google Scholar]
- Smith, S.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008; p. 768. [Google Scholar]
- Karst, J.; Erbilgin, N.; Pec, G.J.; Cigan, P.W.; Najar, A.; Simard, S.W.; Cahill, J.F. Ectomycorrhizal fungi mediate indirect effects of a bark beetle outbreak on secondary chemistry and establishment of pine seedlings. N. Phytol. 2015, 208, 904–914. [Google Scholar] [CrossRef]
- Baath, E. Soil fungal biomass after clear-cutting of a pine forest in central Sweden. Soil Biol. Biochem. 1980, 12, 495–500. [Google Scholar] [CrossRef]
- Newman, S.E.; Davies, F.T.J. High root-zone temperatures mycorrhizal fungi water relations and root hydraulic conductivity of container-grown woody plants. J. Am. Soc. Hortic. Sci. 1988, 113, 138–146. [Google Scholar] [CrossRef]
- Boyle, C.D.; Hellenbrand, K.E. Assessment of the effect of mycorrhizal fungi on drought tolerance of conifer seedlings. Can. J. Bot. 1991, 69, 1764–1771. [Google Scholar] [CrossRef]
- Perry, D.A.; Amaranthus, M.P.; Borchers, J.G.; Borchers, S.L.; Brainerd, R.E. Bootstrapping in ecosystems. Bioscience 1989, 39, 230–237. [Google Scholar] [CrossRef]
- Lang, C.; Polle, A. Ectomycorrhizal fungal diversity, tree diversity and root nutrient relations in a mixed Central European forest. Tree Physiol. 2011, 31, 531–538. [Google Scholar] [CrossRef]
- Jonsson, L.; Dahlberg, A.; Nilsson, M.C.; Karen, O.; Zackrisson, O. Continuity of ectomycorrhizal fungi in self-regenerating boreal Pinus sylvestris forests studied by comparing mycobiont diversity on seedlings and mature trees. N. Phytol. 1999, 142, 151–162. [Google Scholar] [CrossRef]
- Dahlberg, A.; Schimmel, J.; Taylor, A.F.S.; Johannesson, H. Post-fire legacy of ectomycorrhizal fungal communities in the Swedish boreal forest in relation to fire severity and logging intensity. Biol. Conserv. 2001, 100, 151–161. [Google Scholar] [CrossRef]
- Urban, A.; Puschenreiter, M.; Strauss, J.; Gorfer, M. Diversity and structure of ectomycorrhizal and co-associated fungal communities in a serpentine soil. Mycorrhiza 2008, 18, 339–354. [Google Scholar] [CrossRef]
- Rudawska, M.; Wilgan, R.; Janowski, D.; Iwanski, M.; Leski, T. Shifts in taxonomical and functional structure of ectomycorrhizal fungal community of Scots pine (Pinus sylvestris L.) underpinned by partner tree ageing. Pedobiologia 2018, 71, 20–30. [Google Scholar] [CrossRef]
- Aučina, A.; Rudawska, M.; Wilgan, R.; Janowski, D.; Skridaila, A.; Dapkuniene, S.; Leski, T. Functional diversity of ectomycorrhizal fungal communities along a peatland–forest gradient. Pedobiologia 2019, 74, 15–23. [Google Scholar] [CrossRef]
- Colpaert, J.V. Thelephora. In Ectomycorrhizal Fungi. Key Genera in Profile; Cairney, J.W.G., Chambers, S.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 245–325. [Google Scholar]
- Bzdyk, R.M.; Sikora, K.; Studnicki, M.; Aleksandrowicz-Trzcinska, M. Communities of Mycorrhizal Fungi among Seedlings of Scots Pine (Pinus sylvestris L.) Growing on a Clearcut in Microsites Generated by Different Site-Preparation Methods. Forests 2022, 13, 353. [Google Scholar] [CrossRef]
- Kropp, B.R.; Langlois, C.G. Ectomycorrhizae in reforestation. Can. J. For. Res. 1990, 20, 438–451. [Google Scholar] [CrossRef]
- Grebenc, T.; Christensen, M.; Vilhar, U.; Čater, M.; Martin, M.P.; Simončič, P.; Kraigher, H. Response of ectomycorrhizal community structure to gap opening in natural and managed temperate beech-dominated forests. Can. J. For. Res. 2009, 39, 1375–1386. [Google Scholar] [CrossRef]
- Varenius, K.; Karen, O.; Lindahl, B.; Dahlberg, A. Long-term effects of tree harvesting on ectomycorrhizal fungal communities in boreal Scots pine forests. For. Ecol. Manag. 2016, 380, 41–49. [Google Scholar] [CrossRef]
- Simard, S.W.; Durall, D.M. Mycorrhizal networks: A review of their extent, function, and importance. Can. J. Bot. 2004, 82, 1140–1165. [Google Scholar] [CrossRef]
- Douhan, G.W.; Vincenot, L.; Gryta, H.; Selosse, M.A. Population genetics of ectomycorrhizal fungi: From current knowledge to emerging directions. Fungal Biol. 2011, 115, 569–597. [Google Scholar] [CrossRef]
- Varenius, K.; Lindahl, B.D.; Dahlberg, A. Retention of seed trees fails to lifeboat ectomycorrhizal fungal diversity in harvested Scots pine forests. FEMS Microbiol. Ecol. 2017, 93, 1–11. [Google Scholar] [CrossRef]
- Teste, F.P.; Simard, S.W.; Durall, D.M. Role of mycorrhizal networks and tree proximity in ectomycorrhizal colonization of planted seedlings. Fungal Ecol. 2009, 2, 21–30. [Google Scholar] [CrossRef]


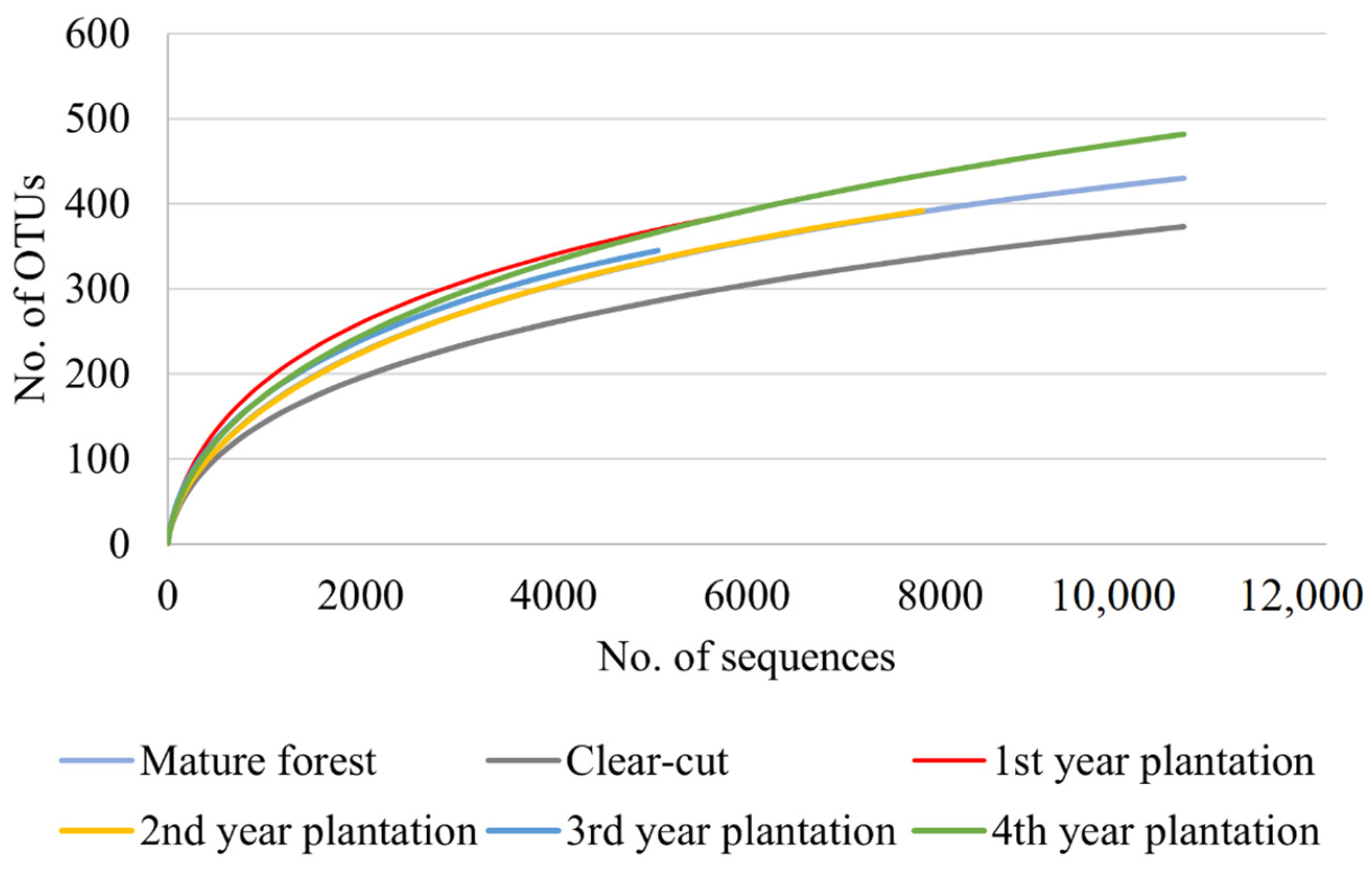


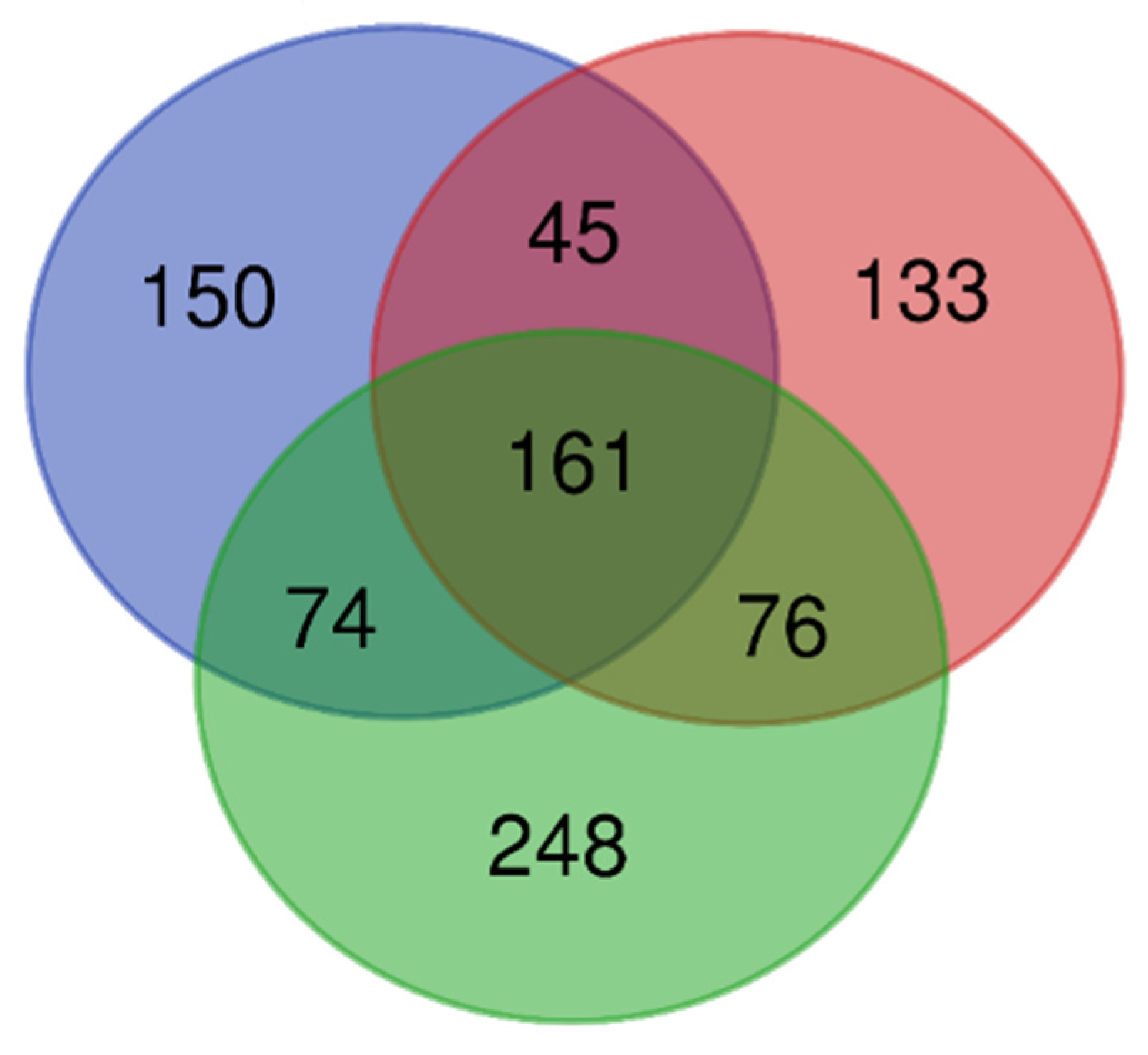
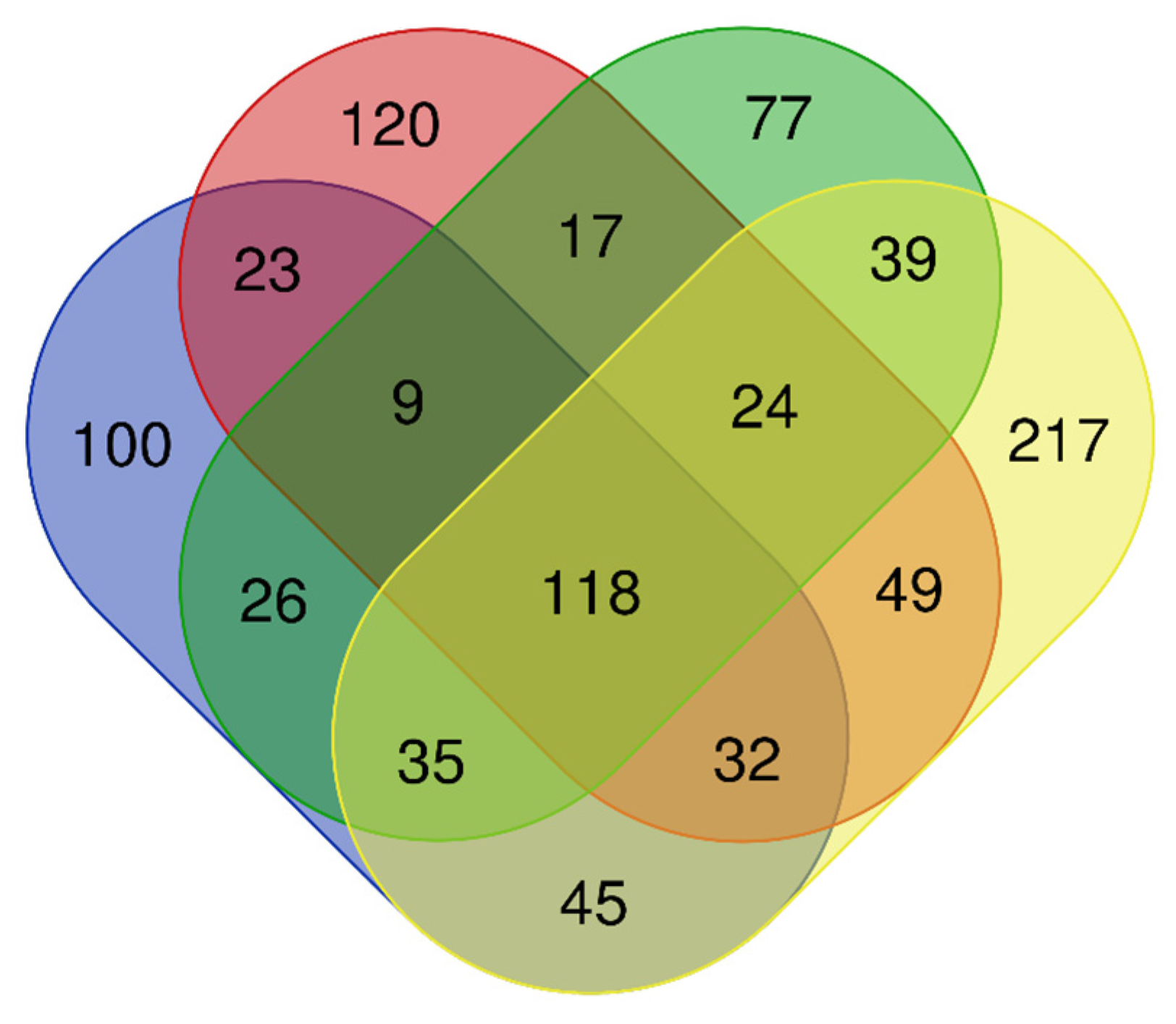


| Region (Code) | Sampling Site Code * | Site Description | Geographic Coordinates | Forest Vegetation Type ** | Stand Age (years) | Mean Tree DBH *** (cm) | Mean Tree Height (m) |
|---|---|---|---|---|---|---|---|
| Alytus (A) | A1 | Mature forest | 54°31′29.4″ N, 24°0′52.9″ E | vm | 94 | 35.9 | 31.2 |
| A2 | Clear-cut | 54°32′33.4″ N, 24°8′22.1″ E | v | - | - | - | |
| A3 | 1st year plantation | 54°34′13.3″ N, 23°57′59.2″ E | vm | 3 | - | - | |
| A4 | 2nd year plantation | 54°31′31.3″ N, 24°0′50.1″ E | vm | 4 | - | - | |
| A5 | 3rd year plantation | 54°31′26.1″ N, 23°56′2.6″ E | vm | 5 | - | - | |
| A6 | 4th year plantation | 54°30′39.2″ N, 23°58′25.0″ E | vm | 6 | - | - | |
| Varėna (V) | V1 | Mature forest | 54°10′45.2″ N, 24°35′9.8″ E | vm | 85 | 28.1 | 24.4 |
| V2 | Clear-cut | 54°8′32.9″ N, 24°34′36.1″ E | v | - | - | - | |
| V3 | 1st year plantation | 54°11′51.9″ N, 24°35′48.7″ E | vm | 3 | - | - | |
| V4 | 2nd year plantation | 54°9′18.4″ N, 24°31′48.3″ E | vm | 4 | - | - | |
| V5 | 3rd year plantation | 54°10′18.7″ N, 24°35′48.9″ E | vm | 5 | - | - | |
| V6 | 4th year plantation | 54°6′7.5″ N, 24°34′4.8″ E | vm | 6 | - | - | |
| Labanoras (L) | L1 | Mature forest | 55°10′36.7″ N, 25°45′28.3″ E | vm | 132 | 36.1 | 31.1 |
| L2 | Clear-cut | 55°9′10.8″ N, 25°47′42.8″ E | v | - | - | - | |
| L3 | 1st year plantation | 55°10′35.0″ N, 25°45′11.5″ E | vm | 3 | - | - | |
| L4 | 2nd year plantation | 55°8′35.9″ N, 25°48′37.0″ E | vm | 4 | - | - | |
| L5 | 3rd year plantation | 55°8′17.9″ N, 25°45′28.3″ E | vm | 5 | - | - | |
| L6 | 4th year plantation | 55°10′37.7″ N, 25°45′33.4″ E | vm | 6 | - | - |
| Site Description | EC | Total P | Total K | Total Ca | Total Mg |
|---|---|---|---|---|---|
| (mS m–1) | (mg kg–1) | ||||
| Mature forest | 2.5 ± 0.1 ab | 195.6 ± 19.1 a | 146.3 ± 8.3 a | 186.7 ± 27.3 a | 242.4 ± 33.8 a |
| Clear-cut | 2.6 ± 0.2 ab | 232.3 ± 29.4 ab | 137.2 ± 7.9 a | 151.2 ± 9.6 a | 229.0 ± 27.8 a |
| 1st year plantation | 3.6 ± 0.5 b | 302.8 ± 33.9 b | 145.4 ± 12.7 a | 191.5 ± 29.6 a | 228.8 ± 40.0 a |
| 2nd year plantation | 2.6 ± 0.2 ab | 250.4 ± 14.5 ab | 122.6 ± 8.8 a | 179.7 ± 20.2 a | 248.2 ± 48.5 a |
| 3rd year plantation | 2.3 ± 0.1 a | 231.9 ± 20.1 ab | 138.6 ± 9.3 a | 206.2 ± 19.4 a | 242.3 ± 41.1 a |
| 4th year plantation | 2.9 ± 0.3 ab | 205.8 ± 11.9 a | 152.0 ± 6.1 a | 211.1 ± 21.0 a | 263.3 ± 24.7 a |
| Cl− | P2O5 | K2O | Ca2+ | Mg2+ | |
| (mg kg–1) | |||||
| Mature forest | 5.8 ± 0.2 a | 79.7 ± 9.2 a | 34.1 ± 2.9 a | 132.6 ± 10.8 ab | 41.8 ± 1.5 a |
| Clear-cut | 6.1 ± 0.4 a | 122.6 ± 26.2 ab | 35.3 ± 4.0 a | 121.3 ± 6.1 ab | 39.9 ± 2.1 a |
| 1st year plantation | 6.7 ± 0.3 a | 169.1 ± 27.8 b | 36.6 ± 4.3 a | 122.8 ± 12.5 ab | 40.8 ± 3.1 a |
| 2nd year plantation | 6.7 ± 0.4 a | 109.8 ± 12.1 ab | 31.3 ± 4.2 a | 120.4 ± 10.8 a | 37.7 ± 2.9 a |
| 3rd year plantation | 6.7 ± 0.4 a | 101.0 ± 11.4 ab | 27.2 ± 2.4 a | 161.6 ± 9.7 b | 48.3 ± 5.6 a |
| 4th year plantation | 6.9 ± 0.4 a | 107.9 ± 16.5 ab | 32.1 ± 3.8 a | 140.8 ± 12.3 ab | 50.6 ± 6.7 a |
| Site Description | No. of Fungal Sequences | No. of Fungal Taxa | Shannon Diversity Index H |
|---|---|---|---|
| Mature forest | 10,523 | 430 | 3.86 |
| Clear-cut | 14,534 | 415 | 3.76 |
| 1st year plantation | 5846 | 388 | 4.30 |
| 2nd year plantation | 7818 | 392 | 4.00 |
| 3rd year plantation | 5078 | 345 | 4.30 |
| 4th year plantation | 17,099 | 559 | 4.21 |
| Total | 60,898 | 1143 |
| OTU | Phylum a | References | Similarity, (%) b | Relative Abundance (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mature Forest | Clear-Cut | 1st Year Plantation | 2nd Year Plantation | 3rd Year Plantation | 4th Year Plantation | Total | ||||
| Archaeorhizomyces sp. 5425_1 | A | MH248043 | 207/207 (100) | 21.59 b | 18.52 ab | 5.03 ab | 0.97 a | 2.40 ab | 9.01 ab | 11.49 |
| Helotiaceae sp. 5425_2 | A | MK131613 | 240/240 (100) | 1.84 a | 9.67 a | 2.51 a | 12.13 a | 5.45 a | 6.91 a | 6.82 |
| Unidentified sp. 5425_18 | A | MN902668 | 258/258 (100) | 1.22 a | 4.67 a | 1.54 a | 3.61 a | 10.73 a | 8.30 a | 5.16 |
| Oidiodendron chlamydosporicum | A | MG597466 | 235/235 (100) | 2.26 ab | 6.31 b | 1.66 ab | 1.06 a | 5.71 ab | 7.94 b | 4.90 |
| Penicillium camemberti | A | MT355566 | 251/251 (100) | 3.24 a | 2.51 a | 2.10 a | 5.95 a | 6.64 a | 3.54 a | 3.67 |
| Cladosporium sp. 5425_16 | A | MT645945 | 243/243 (100) | 4.68 a | 1.42 a | 5.61 a | 2.88 a | 4.21 a | 1.53 a | 2.84 |
| Amanita fulva | B | MT229863 | 260/260 (100) | 2.14 a | 7.08 a | - | - | - | - | 2.06 |
| Jackrogersella multiformis | A | MT573483 | 254/254 (100) | 9.52 a | 0.05 a | 1.54 a | 0.06 a | 1.93 a | 0.05 a | 1.99 |
| Piloderma sphaerosporum | B | MK131527 | 292/292 (100) | 1.45 a | 6.75 a | - | 0.01 a | 0.10 a | 0.01 a | 1.88 |
| Placynthiella oligotropha | A | MK812378 | 248/248 (100) | 0.02 a | - | 0.10 a | 11.61 a | 0.10 a | 1.08 a | 1.81 |
| Malassezia sp. 5425_17 | B | MT594863 | 369/369 (100) | 4.68 b | 0.96 ab | 2.82 ab | 0.84 a | 1.26 ab | 0.87 ab | 1.77 |
| Unidentified sp. 5425_68 | A | EU292507 | 216/217 (99) | 0.29 ab | - | 0.26 ab | 0.43 ab | 0.10 a | 5.22 b | 1.60 |
| Mycena zephirus | B | MH856339 | 304/304 (100) | - | - | 16.08 | - | - | - | 1.54 |
| Phanerochaete sp. 5425_7 | B | KP135164 | 293/293 (100) | 0.45 a | 0.94 ab | 6.55 b | 1.28 ab | 2.38 ab | 0.56 ab | 1.45 |
| Cladophialophora minutissima | B | MT644883 | 313/313 (100) | 0.11 a | 0.21 a | 0.46 a | 8.14 a | 0.20 a | 0.97 a | 1.45 |
| Thelephora terrestris | A | MG597448 | 253/254 (99) | 0.01 ab | 0.01 a | 0.65 ab | - | 1.50 ab | 4.49 b | 1.45 |
| Mortierella sp. 5425_53 | M | HQ021936 | 342/344 (99) | 1.59 a | 0.39 a | 0.33 a | 0.93 a | 2.58 a | 1.39 a | 1.12 |
| Russula decolorans | B | MN992510 | 359/359 (100) | 3.79 b | 1.73 ab | 0.03 a | - | - | 0.05 ab | 1.08 |
| Unidentified sp. 5425_0 | A | JQ312914 | 240/241 (99) | 0.68 a | 1.22 a | 1.03 a | 0.91 a | 1.44 a | 1.16 a | 1.07 |
| Russula vinosa | B | MK838342 | 366/367 (99) | - | - | 0.02 a | - | - | 3.63 a | 1.02 |
| Collophora sp. 5425_14 | A | MW449039 | 244/244 (100) | 1.28 a | 0.72 a | 2.28 a | 0.69 a | 1.81 a | 0.45 a | 0.98 |
| Xylaria hypoxylon | A | MK577428 | 252/252 (100) | 4.41 b | 0.03 ab | 0.51 ab | 0.04 a | 0.85 ab | 0.04 ab | 0.90 |
| Alternaria infectoria | A | MT635276 | 253/253 (100) | 0.29 a | 0.24 a | 1.80 a | 0.52 a | 2.09 a | 1.35 a | 0.90 |
| Mortierella sp. 5425_47 | M | MT601876 | 293/293 (100) | 1.37 b | 0.50 ab | 0.19 a | 0.23 ab | 2.97 ab | 0.34 ab | 0.75 |
| Cladophialophora sp. 5425_111 | A | AB986417 | 268/271 (99) | - | - | - | - | - | 2.54 | 0.71 |
| Mortierella macrocystis | M | MH859487 | 330/330 (100) | 0.45 ab | 1.14 b | 1.25 ab | 0.15 a | 1.34 ab | 0.30 ab | 0.68 |
| Glonium pusillum | A | MT635300 | 249/249 (100) | 0.48 a | 0.43 a | 0.94 a | 1.91 a | 0.51 a | 0.32 a | 0.65 |
| Unidentified sp. 5425_67 | A | HQ433092 | 300/300 (100) | 0.13 ab | 1.86 b | 0.02 a | 0.18 ab | 0.02 a | 0.51 ab | 0.64 |
| Solicoccozyma terricola | B | MH487580 | 329/329 (100) | 0.22 a | 0.52 a | 0.21 a | 1.04 a | 0.20 a | 1.06 a | 0.63 |
| Lecanoromycetes sp. 5425_115 | A | MT027951 | 234/234 (100) | 0.07 a | 0.09 a | 0.31 a | 0.01 a | 0.96 a | 1.66 a | 0.61 |
| Total of OTUs | 68.25 | 67.96 | 55.82 | 55.59 | 57.48 | 65.26 | 63.63 | |||
| OTU | Phy- lum a | References | Similarity, (%) b | Relative Abundance (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mature Forest | Clear-Cut | 1st Year Plantation | 2nd Year Plantation | 3rd Year Plantation | 4th Year Plantation | Total | ||||
| Amanita fulva | B | MT229863 | 260/260 (100) | 2.14 a | 7.08 a | - | - | - | - | 2.06 |
| Piloderma sphaerosporum | B | MK131527 | 292/292 (100) | 1.45 a | 6.75 a | - | 0.01 a | 0.10 a | 0.01 a | 1.88 |
| Thelephora terrestris | B | MT644883 | 313/313 (100) | 0.01 ab | 0.01 a | 0.65 ab | - | 1.50 ab | 4.49 b | 1.45 |
| Russula decolorans | B | MN992510 | 359/359 (100) | 3.79 b | 1.73 ab | 0.03 b | - | - | 0.05 ab | 1.08 |
| Russula vinosa | B | MK838342 | 366/367 (99) | - | - | 0.02 a | - | - | 3.63 a | 1.02 |
| Pseudotomentella sp. 5425_119 | B | HM146848 | 345/345 (100) | - | 2.46 | - | - | - | - | 0.59 |
| Elaphomyces cf. muricatus | A | KR029741 | 287/288 (99) | - | 2.20 | - | - | - | - | 0.53 |
| Amphinema sp. 5425_155 | B | MK838190 | 288/288 (100) | 1.43 a | 0.01 a | - | 0.04 a | - | - | 0.25 |
| Laccaria proxima | B | MN663149 | 301/301 (100) | - | - | 0.43 a | - | 1.75 a | 0.08 a | 0.21 |
| Tricholoma portentosum | B | LC375763 | 304/304 (100) | 0.1a | 0.65 a | - | 0.01 a | 0.02 a | - | 0.19 |
| Amanita pantherina | B | MZ410633 | 310/310 (100) | 0.13 a | - | - | - | - | 0.59 a | 0.19 |
| Russula vesca | B | MZ410735 | 320/321 (99) | 0.76 a | 0.13 a | - | 0.04 a | - | - | 0.17 |
| Russula albonigra | B | DQ422029 | 321/321 (100) | 0.95 | - | - | - | - | - | 0.16 |
| Russula adusta | B | MG687346 | 321/321 (100) | - | 0.65 a | 0.07 a | - | - | - | 0.16 |
| Piloderma bicolor | B | MH809947 | 283/283 (100) | - | 0.65 | - | - | - | - | 0.15 |
| Xerocomus badius | B | MN947388 | 284/284 (100) | 0.04 a | - | 0.03 a | - | - | 0.46 a | 0.14 |
| Russula paludosa | B | LC192779 | 358/358 (100) | - | 0.44 a | 0.02 a | - | - | - | 0.11 |
| Pseudotomentella sp. 5425_255 | B | MK290703 | 316/316 (100) | - | 0.36 | - | - | - | - | 0.09 |
| Tomentella stuposa | B | MK838269 | 313/314 (99) | - | 0.30 | - | - | - | - | 0.07 |
| Tricholoma virgatum | B | DQ389735 | 302/302 (100) | - | 0.30 | - | - | - | - | 0.07 |
| Amanita porphyria | B | MT345267 | 296/296 (100) | 0.01 a | 0.21 a | 0.02 a | - | - | - | 0.05 |
| Cortinarius sp. 5425_463 | B | MG597372 | 303/305 (99) | 0.28 | - | - | - | - | - | 0.05 |
| Wilcoxina sp. 5425_54 | A | MW238026 | 257/257 (100) | - | 0.01 a | - | - | - | 0.11 a | 0.03 |
| Pseudotomentella griseopergamacea | B | MG597410 | 321/321 (100) | - | - | - | - | - | 0.11 | 0.03 |
| Russula aeruginea | B | MG680182 | 301/301 (100) | 0.13 a | 0.01 a | - | 0.04 a | - | - | 0.03 |
| Albatrellus sp. 5425_551 | B | HQ204631 | 330/338 (98) | - | 0.12 | - | - | - | - | 0.03 |
| Inocybe sp. 5425_176 | B | MK838270 | 303/303 (100) | - | - | - | - | - | 0.10 | 0.03 |
| Piloderma olivaceum | B | MH864982 | 281/281 (100) | 0.14 a | 0.01 a | - | - | - | - | 0.03 |
| Inocybe lutescens | B | MW845894 | 295/295 (100) | 0.13 a | 0.01 a | - | - | - | 0.01a | 0.03 |
| Tylospora fibrillosa | B | MF926576 | 287/287 (100) | - | 0.10 | - | - | - | - | 0.02 |
| Total of OTUs | 11.57 | 24.17 | 1.27 | 0.14 | 3.37 | 9.63 | 10.89 | |||
| Variables | pHCaCl2 | SOC | N | P | K | Ca | Mg | Ca2+ | Mg2+ | Cl− |
|---|---|---|---|---|---|---|---|---|---|---|
| g kg−1 | mg kg−1 | |||||||||
| No. of fungal sequences | 0.050 | 0.041 | 0.114 | −0.342 | 0.262 | 0.099 | 0.327 | −0.079 | −0.153 | 0.493 |
| No. of fungal OTUs | −0.212 | 0.321 | 0.336 | 0.011 | 0.324 | 0.299 | 0.303 | 0.152 | 0.110 | 0.483 |
| No. of mycorrhizal fungi | −0.413 | 0.518 * | 0.498 * | −0.177 | 0.477 | 0.225 | 0.479 | 0.212 | 0.066 | 0.021 |
| No. of pathogenic fungi | −0.028 | 0.064 | 0.087 | 0.263 | −0.085 | 0.078 | −0.120 | −0.110 | −0.181 | 0.389 |
| No. of saprotrophic fungi | −0.173 | 0.271 | 0.350 | 0.262 | 0.406 | 0.493 | 0.298 | 0.477 | 0.505 * | 0.206 |
| No. of endophytes | −0.207 | 0.436 | 0.380 | 0.141 | 0.435 | 0.142 | 0.384 | 0.068 | 0.158 | 0.245 |
| Shannon_H | −0.274 | 0.183 | 0.003 | 0.363 | −0.090 | −0.099 | −0.200 | 0.192 | 0.383 | −0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Povilaitienė, A.; Gedminas, A.; Varnagirytė-Kabašinskienė, I.; Marčiulynienė, D.; Marčiulynas, A.; Lynikienė, J.; Mishcherikova, V.; Menkis, A. Changes in Chemical Properties and Fungal Communities of Mineral Soil after Clear-Cutting and Reforestation of Scots Pine (Pinus sylvestris L.) Sites. Forests 2022, 13, 1780. https://doi.org/10.3390/f13111780
Povilaitienė A, Gedminas A, Varnagirytė-Kabašinskienė I, Marčiulynienė D, Marčiulynas A, Lynikienė J, Mishcherikova V, Menkis A. Changes in Chemical Properties and Fungal Communities of Mineral Soil after Clear-Cutting and Reforestation of Scots Pine (Pinus sylvestris L.) Sites. Forests. 2022; 13(11):1780. https://doi.org/10.3390/f13111780
Chicago/Turabian StylePovilaitienė, Aistė, Artūras Gedminas, Iveta Varnagirytė-Kabašinskienė, Diana Marčiulynienė, Adas Marčiulynas, Jūratė Lynikienė, Valeriia Mishcherikova, and Audrius Menkis. 2022. "Changes in Chemical Properties and Fungal Communities of Mineral Soil after Clear-Cutting and Reforestation of Scots Pine (Pinus sylvestris L.) Sites" Forests 13, no. 11: 1780. https://doi.org/10.3390/f13111780





