Effects of Intercropping Pandanus amaryllifolius on Soil Properties and Microbial Community Composition in Areca Catechu Plantations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Experimental Design and Management
2.3. Soil Sampling
2.4. Analysis of Soil Physicochemical Properties and Soil Enzyme Activities
2.5. Soil DNA Extraction and Sequencing
2.6. Bioinformatics Analysis
2.7. Statistical Analysis
3. Results
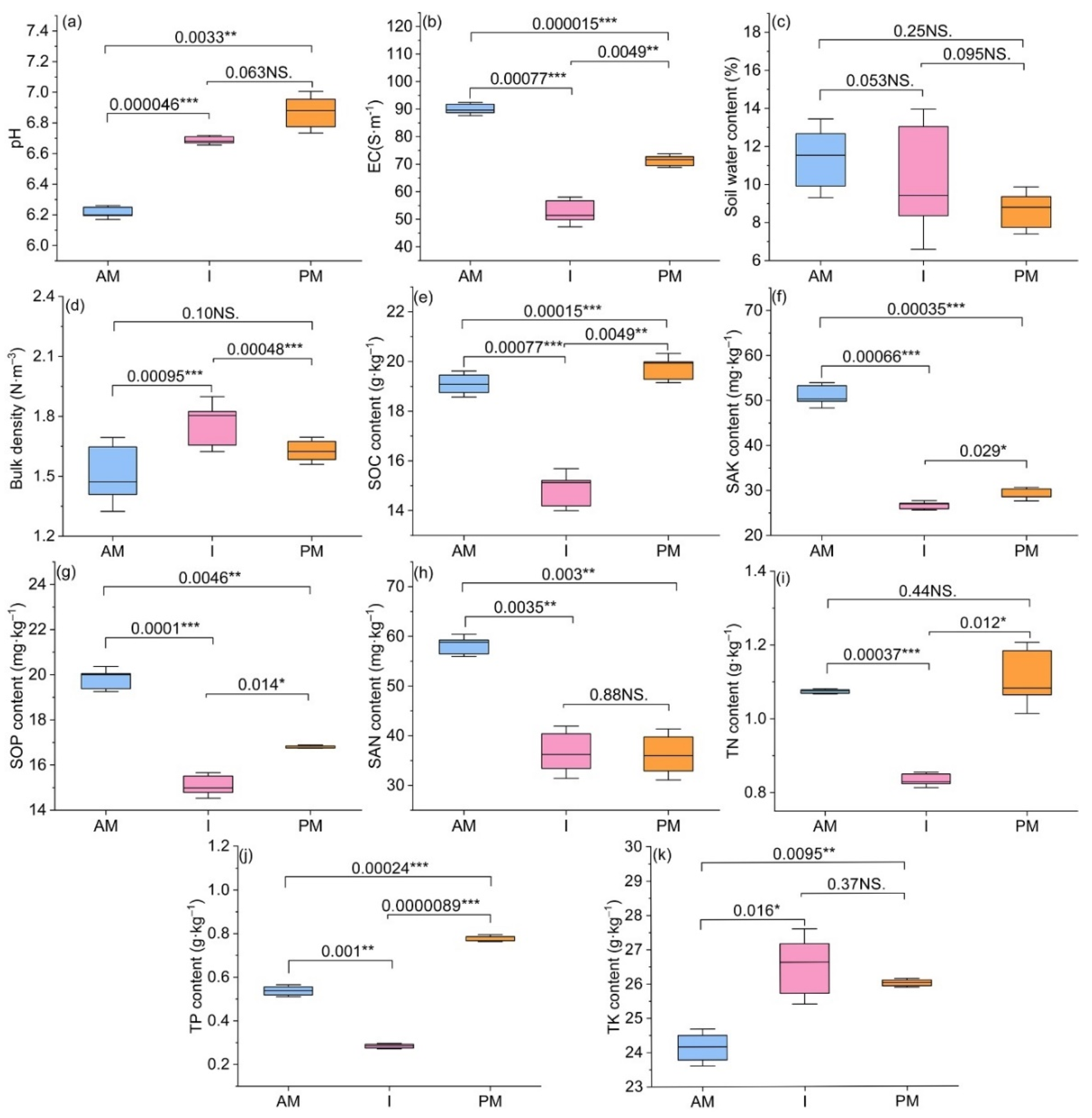
3.1. Changes in Soil Physicochemical Properties and Enzyme Activities
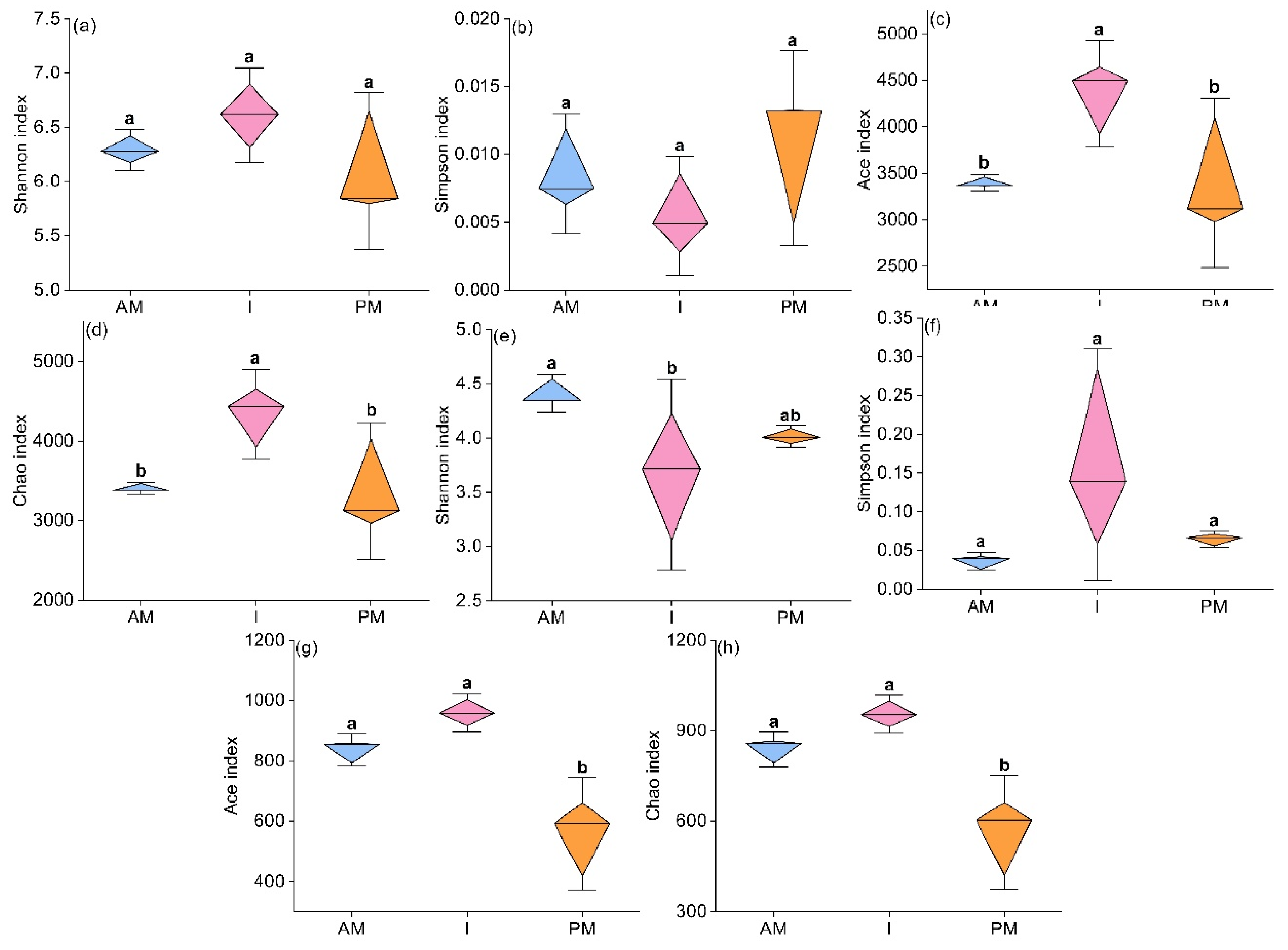
3.2. Changes in Soil Microbial Community Diversity
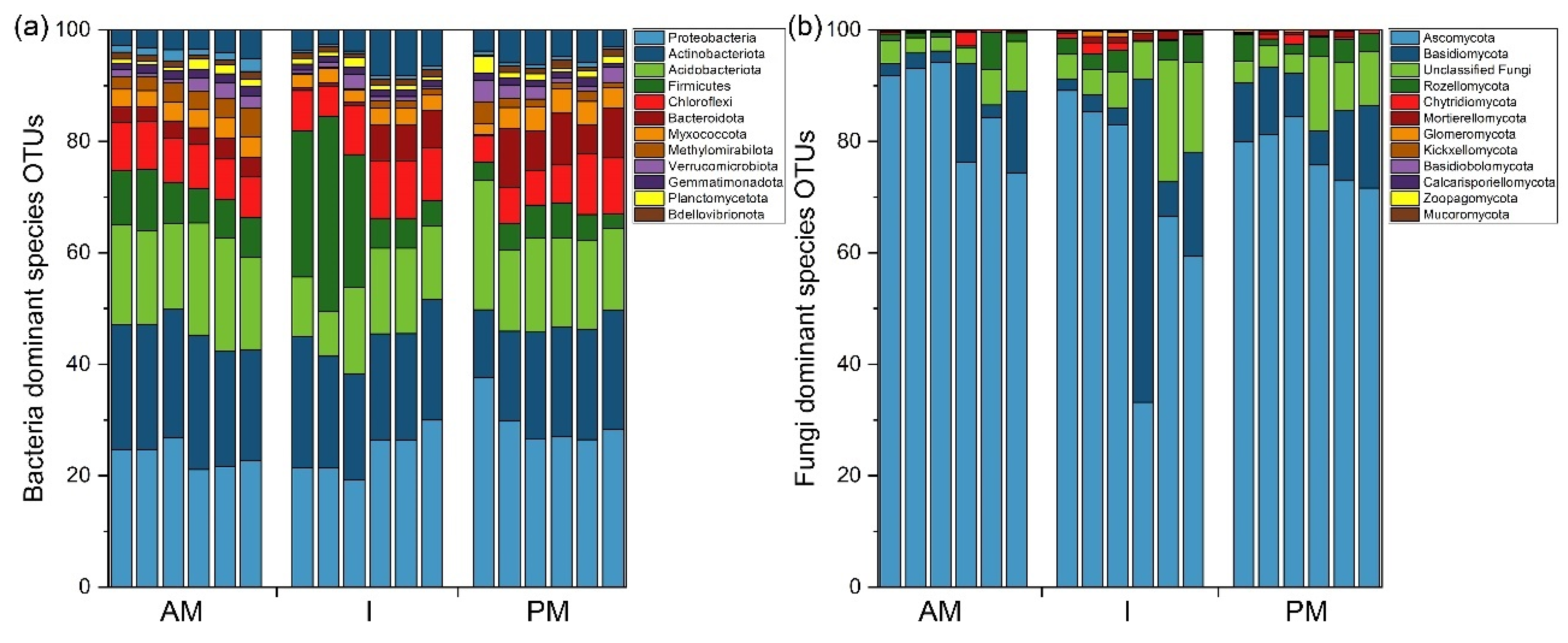
3.3. Changes in the Composition and Structure of Soil Microbial Community
3.4. Changes in Soil Microbial Functional Profiles
3.5. Relationship between Soil Properties and Soil Enzymes
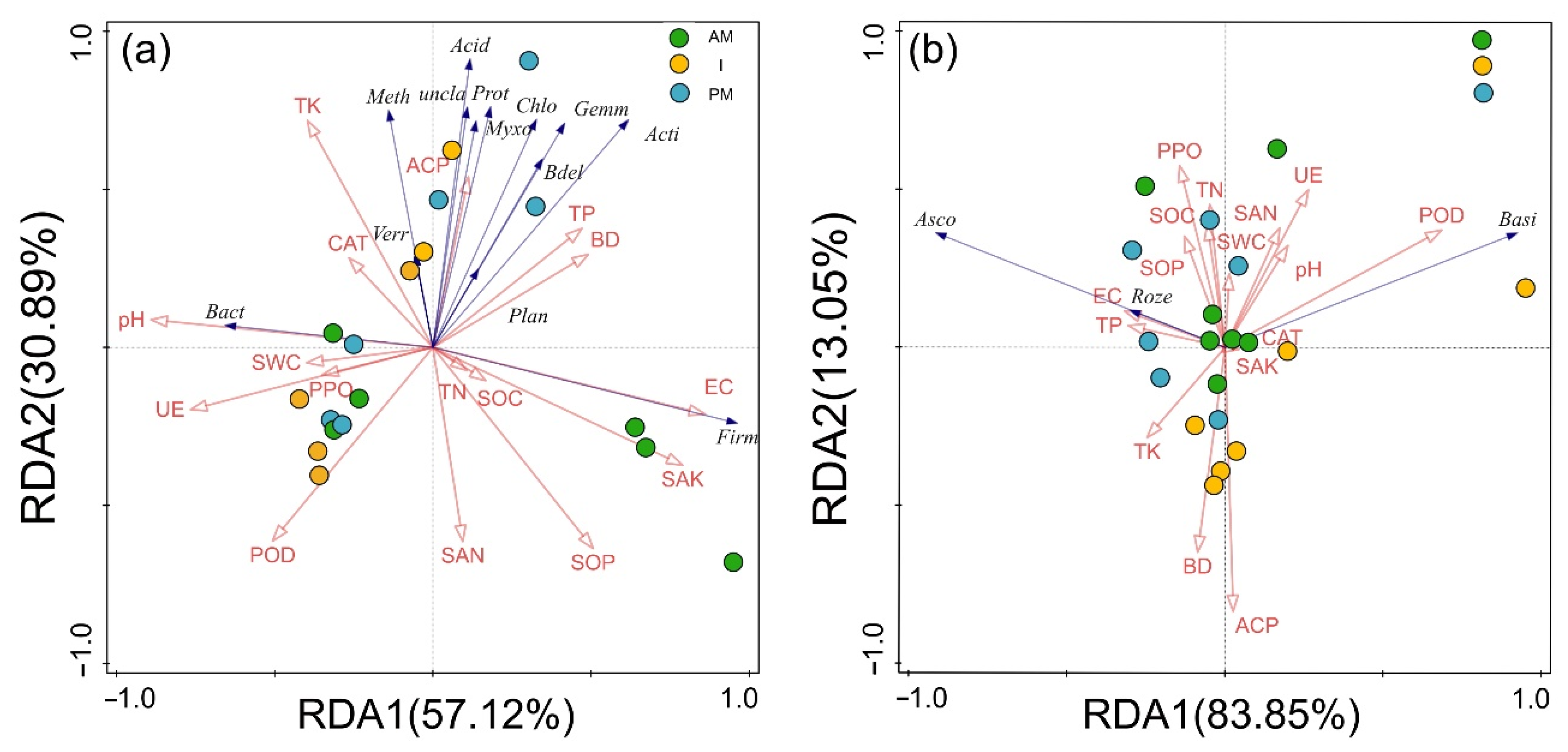
3.6. Influence of Soil Biological and Abiotic Factors on Soil Microbial Community Diversity
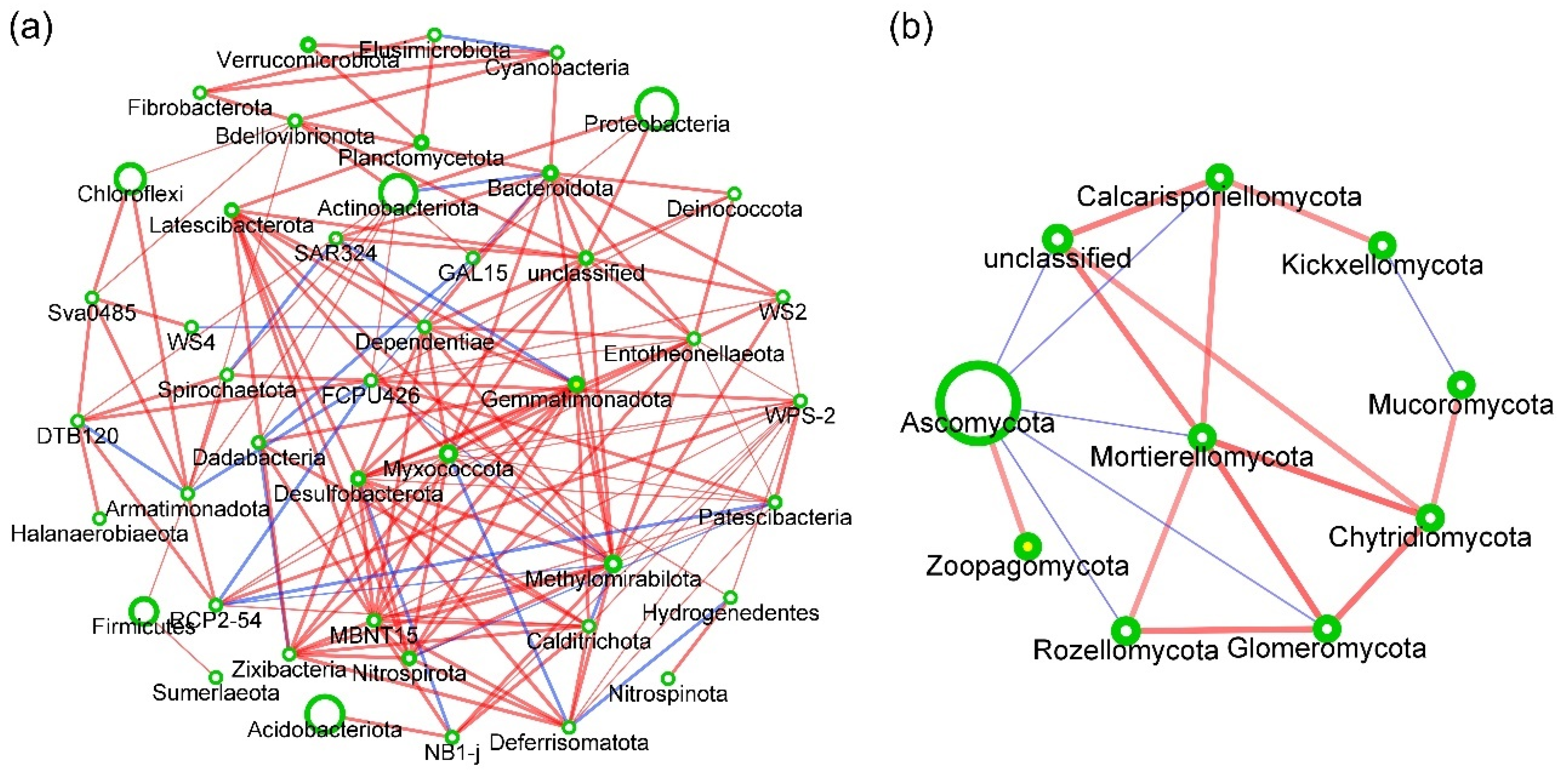
3.7. Responses of Soil Microbial Community Structure to Three Planting Modes
4. Discussion
4.1. Effects of Planting Modes on Soil Enzyme Activities
4.2. Regulatory Mechanisms of Planting Modes on Soil Microbial Diversity
4.3. Key Regulatory Factors of Different Planting Modes on Soil Microbial Community Composition and Structure
4.4. Effects of Different Planting Modes on Soil Microbial Functional Groups
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| PM | I | AM | |
|---|---|---|---|
| Bacteria | |||
| Proteobacteria | 19,202.33 ± 332.93 b | 20,230.33 ± 1974.88 b | 24,117.33 ± 866.37 a |
| Actinobacteriota | 16,801.00 ± 1021.31 b | 15,365.33 ± 712.89 b | 20,129.67 ± 298.15 a |
| Acidobacteriota | 10,725.33 ± 1697.53 a | 14,804.67 ± 2381.47 a | 13,546.33 ± 2433.41 a |
| Firmicutes | 14,715.33 ± 3308.49 a | 4676.67 ± 519.08 b | 6590.33 ± 1202.16 b |
| Chloroflexi | 6552.67 ± 992.70 ab | 5415.00 ± 93.15 b | 7745.33 ± 1054.78 a |
| Bacteroidota | 3069.67 ± 894.62 a | 3479.67 ± 1728.98 a | 4053.67 ± 167.99 a |
| Myxococcota | 2568.67 ± 160.48 a | 2770.00 ± 427.36 a | 2750.33 ± 315.78 a |
| Methylomirabilota | 458.33 ± 176.98 c | 2624.33 ± 531.17 a | 1812.00 ± 317.02 b |
| Verrucomicrobiota | 1187.67 ± 936.47 ab | 2081.67 ± 328.36 a | 706.33 ± 296.01 c |
| Gemmatimonadota | 920.33 ± 126.75 a | 1126.67 ± 29.30 a | 1074.33 ± 122.40 a |
| Planctomycetota | 915.67 ± 307.42 a | 1308.33 ± 496.08 a | 602.33 ± 101.11 a |
| Bdellovibrionota | 811.00 ± 149.08 a | 751.67 ± 255.58 a | 1102.67 ± 31.13 a |
| unclassified Bacteria | 420.00 ± 51.68 b | 909.67 ± 280.23 a | 994.33 ± 206.93 a |
| Fungi | |||
| Ascomycota | 107,293.00 ± 6816.14 a | 85,814.67 ± 10,179.97 b | 112,730.67 ± 5197.99 a |
| Basidiomycota | 14,402.00 ± 2356.95 a | 20,973.67 ± 18,341.30 a | 9764.33 ± 5609.52 a |
| Unclassified Fungi | 10,493.67 ± 3031.47 a | 13,498.33 ± 5525.81 a | 6377.67 ± 2092.01 a |
| Rozellomycota | 4314.00 ± 1480.56 a | 3867.67 ± 2074.86 a | 2791.00 ± 2524.00 a |
| Bacteria | PM | I | AM |
|---|---|---|---|
| Aerobic_chemoheterotrophy | 8183.83 ± 1148.68 a | 5707.67 ± 1096.84 b | 6216.83 ± 1358.33 b |
| Chemoheterotrophy | 7349.50 ± 3765.68 a | 5907.00 ± 1153.43 a | 6550.17 ± 1440.54 a |
| Animal_parasites_or_symbionts | 1519.00 ± 1102.83 a | 913.33 ± 291.12 a | 760.67 ± 135.74 a |
| Human_pathogens_all | 1490.5 ± 1089.56 a | 892.83 ± 301.78 a | 721.50 ± 146.22 a |
| Nitrate_reduction | 1326.00 ± 944.36 a | 735.67 ± 270.20 a | 677.83 ± 207.45 a |
| Nitrogen_fixation | 590.17 ± 316.09 b | 726.67 ± 91.09 ab | 897.17 ± 201.86 a |
| Predatory_or_exoparasitic | 477.67 ± 291.51 a | 504.67 ± 170.19 a | 500.17 ± 73.51 a |
| Human_pathogens_pneumonia | 538.50 ± 276.01 a | 538.33 ± 119.02 a | 398.67 ± 162.01 a |
| Invertebrate_parasites | 882.33 ± 887.74 a | 292.83 ± 211.82 a | 281.83 ± 157.54 a |
| Aromatic_compound_degradation | 561.50 ± 166.59 b | 401.33 ± 208.33 ab | 227.17 ± 104.81 a |
| Chitinolysis | 636.83 ± 657.41 a | 289.83 ± 315.06 a | 155.50 ± 148.64 a |
| Ureolysis | 316.00 ± 204.44 a | 337.00 ± 329.12 a | 326.83 ± 137.34 a |
| Phototrophy | 403.33 ± 428.91 a | 277.17 ± 69.48 a | 244.33 ± 51.35 a |
| Cellulolysis | 127.83 ± 71.19 b | 208.50 ± 147.37 ab | 503.67 ± 469.15 a |
| Photoautotrophy | 380.50 ± 428.29 a | 233.17 ± 66.01 a | 172.00 ± 85.81 a |
| Fermentation | 192.83 ± 104.82 a | 195.67 ± 165.37 a | 302.50 ± 152.66 a |
| Others | 2946.83 ± 1281.30 a | 2191.50 ± 614.30 a | 2000.17 ± 1076.32 a |
| Fungi | AM | I | PM |
|---|---|---|---|
| Wood_Saprotroph | 11,270.17 ± 7010.83 a | 4806.83 ± 1352.41 b | 7946.67 ± 1797.94 ab |
| Soil_Saprotroph | 858.17 ± 164.50 a | 417.33 ± 245.91 a | 894.17 ± 827.92 a |
| Plant_Pathogen | 1706.00 ± 565.32 a | 1601.50 ± 2539.11 a | 2047.33 ± 1870.07 a |
| Plant_Pathogen_Wood_Saprotroph | 1186.83 ± 891.84 a | 333.83 ± 267.63 b | 344.83 ± 608.14 b |
| Animal_Pathogen | 453.83 ± 361.55 ab | 689.67 ± 280.65 a | 221.83 ± 242.93 b |
| Endophyte | 16.17 ± 24.31 b | 645.67 ± 699.13 a | 433.17 ± 150.1 ab |
| Ectomycorrhizal | 28.17 ± 20.95 a | 696.00 ± 1028.55 a | 128.17 ± 100.88 a |
| Fungal_Parasite | 93.50 ± 54.34 a | 256.17 ± 315.36 a | 263.17 ± 198.15 a |
| Arbuscular_Mycorrhizal | 8.33 ± 5.47 b | 258.67 ± 239.75 a | 115.33 ± 112.58 ab |
| Epiphyte | 274.67 ± 423.17 a | 0.33 ± 0.82 a | 79.50 ± 193.27 a |
| Dung_Saprotroph | 101.00 ± 47.92 a | 27.00 ± 24.76 a | 134.83 ± 142.33 a |
| Plant_Saprotroph | 55.00 ± 27.40 a | 27.00 ± 21.72 a | 58.00 ± 58.36 a |
| Others | 4.17 ± 1.72 a | 21.67 ± 23.75 a | 36.67 ± 63.59 a |
| Soil Microorganisms | Regression Model | R2 | F Value | p Value |
|---|---|---|---|---|
| Actinobacteriota | Actinobacteriota = 2831.888 + 11,364.551 × TP + 325.103 × TK | 0.951 | 57.843 | 0.000 |
| Ascomycota | Ascomycota = 10,079.723 + 5134.928 × SOC | 0.734 | 19.297 | 0.003 |
| Chloroflexi | Chloroflexi = −906.557 + 7427.543 × TN | 0.643 | 12.632 | 0.009 |
| Firmicutes | Firmicutes = −5205.866 + 388.736 × SAK | 0.846 | 38.479 | 0.000 |
| Methylomirabilota | Methylomirabilota = 5797.752 − 58.409 × EC | 0.908 | 69.196 | 0.000 |
| Proteobacteria | Proteobacteria = 5845.745 + 21.631 × URE | 0.761 | 22.343 | 0.002 |
| Unclassified Bacteria | Unclassified Bacteria = −8218.610 + 1235.298 × pH + 84.372 × SWC | 0.913 | 31.443 | 0.001 |
| Verrucomicrobiota | Verrucomicrobiota = 5945.663 − 258.262 × SOC | 0.575 | 9.469 | 0.018 |
| Name | Explains (%) | F | P |
|---|---|---|---|
| Environment-Bacteria | |||
| pH | 24.5 | 13.6 | 0.002 |
| TP | 48.6 | 6.6 | 0.004 |
| POD | 11.2 | 3.6 | 0.022 |
| EC | 6.4 | 2.7 | 0.108 |
| UE | 3.8 | 2.0 | 0.146 |
| SOC | 2.3 | 1.4 | 0.312 |
| SAK | 1.8 | 1.3 | 0.434 |
| SOP | 1.4 | <0.1 | 1 |
| Environment-Fungi | |||
| SAN | 23.5 | 2.2 | 0.144 |
| TK | 11.6 | 1.1 | 0.330 |
| BD | 28.0 | 3.8 | 0.102 |
| SWC | 8.2 | 1.2 | 0.354 |
| PPO | 18.5 | 5.5 | 0.064 |
| UE | 9.4 | 26.5 | 0.032 |
| POD | 0.3 | 0.7 | 0.554 |
| pH | 0.4 | <0.1 | 1 |

References
- Duchene, O.; Vian, J.; Celette, F. Intercropping with legume for agroecological cropping systems: Complementarity and facilitation processes and the importance of soil microorganisms. A review. Agric. Ecosyst. Environ. 2017, 240, 148–161. [Google Scholar] [CrossRef]
- Cuartero, J.; Pascual, J.A.; Vivo, J.; özbolat, O.; Sánchez-Navarro, V.; Egea-Cortines, M.; Zornoza, R.; Mena, M.M.; Garcia, E.; Ros, M. A first-year melon/cowpea intercropping system improves soil nutrients and changes the soil microbial community. Agric. Ecosyst. Environ. 2022, 328, 107856. [Google Scholar] [CrossRef]
- Kang, Z.; Gong, M.; Li, Y.; Chen, W.; Yang, Y.; Qin, J.; Li, H. Low Cd-accumulating rice intercropping with Sesbania cannabina L. reduces grain Cd while promoting phytoremediation of Cd-contaminated soil. Sci. Total Environ. 2021, 800, 149600. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Heerink, N.; Jin, S.; Berentsen, P.; Zhang, L.; van der Werf, W. Intercropping and agroforestry in China—Current state and trends. Agric. Ecosyst. Environ. 2017, 244, 52–61. [Google Scholar] [CrossRef]
- Nyawade, S.O.; Karanja, N.N.; Gachene, C.K.K.; Gitari, H.I.; Schulte-Geldermann, E.; Parker, M.L. Short-term dynamics of soil organic matter fractions and microbial activity in smallholder potato-legume intercropping systems. Appl. Soil Ecol. 2019, 142, 123–135. [Google Scholar] [CrossRef]
- Gu, C.; Bastiaans, L.; Anten, N.P.R.; Makowski, D.; van der Werf, W. Annual intercropping suppresses weeds: A meta-analysis. Agric. Ecosyst. Environ. 2021, 322, 107658. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; He, Z.; Xing, C.; Zhu, J.; Gu, X.; Lan, Y.; Wu, Z.; Liao, P.; Zhu, D. Forest gaps mediate the structure and function of the soil microbial community in a Castanopsis kawakamii forest. Ecol. Indic. 2021, 122, 107288. [Google Scholar] [CrossRef]
- Bhat, R.; Sujatha, S.; Jose, C.T. Assessing soil fertility of a laterite soil in relation to yield of arecanut (Areca catechu L.) in humid tropics of India. Geoderma 2012, 189-190, 91–97. [Google Scholar] [CrossRef]
- Li, J.; Liu, L.; Zhou, H.; Li, M. Improved Viability of Areca (Areca catechu L.) Seedlings under Drought Stress Using a Superabsorbent Polymer. Hortscience 2018, 53, 1872–1876. [Google Scholar] [CrossRef] [Green Version]
- Santonja, M.; Rancon, A.; Fromin, N.; Baldy, V.; Hättenschwiler, S.; Fernandez, C.; Montès, N.; Mirleau, P. Plant litter diversity increases microbial abundance, fungal diversity, and carbon and nitrogen cycling in a Mediterranean shrubland. Soil Biol. Biochem. 2017, 111, 124–134. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Jaafar, H.Z. Profiling of phenolic compounds and their antioxidant and anticancer activities in pandan (Pandanus amaryllifolius Roxb.) extracts from different locations of Malaysia. BMC Complement. Altern. Med. 2013, 13, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quyen, N.T.C.; Quyen, N.T.N.; Nhan, L.T.H.; Toan, T.Q. Antioxidant activity, total phenolics and flavonoids contents of Pandanus amaryllifolius (Roxb.). IOP Conf. Ser. Mater. Sci. Eng. 2020, 991, 12019. [Google Scholar] [CrossRef]
- Routray, W.; Rayaguru, K. Chemical Constituents and Post-Harvest Prospects of Pandanus amaryllifolius Leaves: A Review. Food Rev. Int. 2010, 26, 230–245. [Google Scholar] [CrossRef]
- Veres, Z.; Kotroczó, Z.; Fekete, I.; Tóth, J.A.; Lajtha, K.; Townsend, K.; Tóthmérész, B. Soil extracellular enzyme activities are sensitive indicators of detrital inputs and carbon availability. Appl. Soil Ecol. 2015, 92, 18–23. [Google Scholar] [CrossRef]
- Gong, X.; Liu, C.; Li, J.; Luo, Y.; Yang, Q.; Zhang, W.; Yang, P.; Feng, B. Responses of rhizosphere soil properties, enzyme activities and microbial diversity to intercropping patterns on the Loess Plateau of China. Soil Tillage Res. 2019, 195, 104355. [Google Scholar] [CrossRef]
- Cordero, I.; Snell, H.; Bardgett, R.D. High throughput method for measuring urease activity in soil. Soil Biol. Biochem. 2019, 134, 72–77. [Google Scholar] [CrossRef]
- Lopes, É.M.G.; Reis, M.M.; Frazão, L.A.; Da Mata Terra, L.E.; Lopes, E.F.; Dos Santos, M.M.; Fernandes, L.A. Biochar increases enzyme activity and total microbial quality of soil grown with sugarcane. Environ.Technol. Innov. 2021, 21, 101270. [Google Scholar] [CrossRef]
- Piotrowska-Długosz, A.; Długosz, J.; Frąc, M.; Gryta, A.; Breza-Boruta, B. Enzymatic activity and functional diversity of soil microorganisms along the soil profile—A matter of soil depth and soil-forming processes. Geoderma 2022, 416, 115779. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Leptin, A.; Whitehead, D.; Anderson, C.R.; Cameron, K.C.; Lehto, N.J. Increased soil nitrogen supply enhances root-derived available soil carbon leading to reduced potential nitrification activity. Appl. Soil Ecol. 2021, 159, 103842. [Google Scholar] [CrossRef]
- Vimal, S.R.; Singh, J.S.; Arora, N.K.; Singh, S. Soil-Plant-Microbe Interactions in Stressed Agriculture Management: A Review. Pedosphere 2017, 27, 177–192. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Yang, J.; Duan, Y.; Luo, Y.; Taherzadeh, M.J.; Li, Y.; Li, H.; Awasthi, M.K.; Zhao, Z. The diversity of microbial community and function varied in response to different agricultural residues composting. Sci. Total Environ. 2020, 715, 136983. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gu, H.; Yao, Q.; Jiao, F.; Liu, J.; Jin, J.; Liu, X.; Wang, G. Microbial communities in the diagnostic horizons of agricultural Isohumosols in northeast China reflect their soil classification. Catena 2022, 216, 106430. [Google Scholar] [CrossRef]
- Zhou, X.; Yu, G.; Wu, F. Effects of intercropping cucumber with onion or garlic on soil enzyme activities, microbial communities and cucumber yield. Eur. J. Soil Biol. 2011, 47, 279–287. [Google Scholar] [CrossRef]
- Jing, H.; Li, J.; Yan, B.; Wei, F.; Wang, G.; Liu, G. The effects of nitrogen addition on soil organic carbon decomposition and microbial C-degradation functional genes abundance in a Pinus tabulaeformis forest. Forest Ecol. Manag. 2021, 489, 119098. [Google Scholar] [CrossRef]
- Huang, H.; Tian, D.; Zhou, L.; Su, H.; Ma, S.; Feng, Y.; Tang, Z.; Zhu, J.; Ji, C.; Fang, J. Effects of afforestation on soil microbial diversity and enzyme activity: A meta-analysis. Geoderma 2022, 423, 115961. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Yang, W.; Yang, X.; Li, W.; Xia, Q.; Li, J.; Gao, Z.; Yang, Z. Rhizosphere soil properties, microbial community, and enzyme activities: Short-term responses to partial substitution of chemical fertilizer with organic manure. J. Environ. Manag. 2021, 299, 113650. [Google Scholar] [CrossRef]
- Sekaran, U.; Loya, J.R.; Abagandura, G.O.; Subramanian, S.; Owens, V.; Kumar, S. Intercropping of kura clover (Trifolium ambiguum M. Bieb) with prairie cordgrass (Spartina pectinata link.) enhanced soil biochemical activities and microbial community structure. Appl. Soil Ecol. 2020, 147, 103427. [Google Scholar] [CrossRef]
- Zhu, L.; He, J.; Tian, Y.; Li, X.; Li, Y.; Wang, F.; Qin, K.; Wang, J. Intercropping Wolfberry with Gramineae plants improves productivity and soil quality. Sci. Hortic. 2022, 292, 110632. [Google Scholar] [CrossRef]
- Mathesius, U. Are legumes different? Origins and consequences of evolving nitrogen fixing symbioses. J. Plant Physiol. 2022, 276, 153765. [Google Scholar] [CrossRef]
- Wang, L.; Deng, D.; Feng, Q.; Xu, Z.; Pan, H.; Li, H. Changes in litter input exert divergent effects on the soil microbial community and function in stands of different densities. Sci. Total Environ. 2022, 845, 157297. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Shao, G.; Pang, Y.; Dai, H.; Zhang, L.; Yan, P.; Zou, Z.; Zhang, Z.; Xu, J.; Zamanian, K.; et al. Enhanced soil quality after forest conversion to vegetable cropland and tea plantations has contrasting effects on soil microbial structure and functions. Catena 2022, 211, 106029. [Google Scholar] [CrossRef]
- Gilmullina, A.; Rumpel, C.; Blagodatskaya, E.; Chabbi, A. Management of grasslands by mowing versus grazing – impacts on soil organic matter quality and microbial functioning. Appl. Soil Ecol. 2020, 156, 103701. [Google Scholar] [CrossRef]
- van Agtmaal, M.; Straathof, A.L.; Termorshuizen, A.; Lievens, B.; Hoffland, E.; de Boer, W. Volatile-mediated suppression of plant pathogens is related to soil properties and microbial community composition. Soil Biol. Biochem. 2018, 117, 164–174. [Google Scholar] [CrossRef]
- Ablimit, R.; Li, W.; Zhang, J.; Gao, H.; Zhao, Y.; Cheng, M.; Meng, X.; An, L.; Chen, Y. Altering microbial community for improving soil properties and agricultural sustainability during a 10-year maize-green manure intercropping in Northwest China. J. Environ. Manag. 2022, 321, 115859. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Q.; Zhang, D.; Jia, W.; Chen, J.; Liu, G.; Cheng, X. The ratio of ligninase to cellulase increased with the reduction of plant detritus input in a coniferous forest in subtropical China. Appl. Soil Ecol. 2022, 170, 104269. [Google Scholar] [CrossRef]
- Luo, G.; Sun, B.; Li, L.; Li, M.; Liu, M.; Zhu, Y.; Guo, S.; Ling, N.; Shen, Q. Understanding how long-term organic amendments increase soil phosphatase activities: Insight into phoD- and phoC-harboring functional microbial populations. Soil Biol. Biochem. 2019, 139, 107632. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, J.; Xing, Y.; Xie, X.; Wang, L. Influence of intercropping Chinese milk vetch on the soil microbial community in rhizosphere of rape. Plant Soil 2019, 440, 85–96. [Google Scholar] [CrossRef]
- Li, H.; Qiu, Y.; Yao, T.; Han, D.; Gao, Y.; Zhang, J.; Ma, Y.; Zhang, H.; Yang, X. Nutrients available in the soil regulate the changes of soil microbial community alongside degradation of alpine meadows in the northeast of the Qinghai-Tibet Plateau. Sci. Total Environ. 2021, 792, 148363. [Google Scholar] [CrossRef]
- Wang, H.; Feng, D.; Zhang, A.; Zheng, C.; Li, K.; Ning, S.; Zhang, J.; Sun, C. Effects of saline water mulched drip irrigation on cotton yield and soil quality in the North China Plain. Agric. Water Manag. 2022, 262, 107405. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, L.; Liu, C.; Jin, Y.; Li, F.; Khan, S.; Liang, X. Reduced colloidal phosphorus loss potential and enhanced phosphorus availability by manure-derived biochar addition to paddy soils. Geoderma 2021, 402, 115348. [Google Scholar] [CrossRef]
- Bray, R.H.; Kurtz, L.T. Determination of Total, Organic, and Available Forms of Phosphorus in Soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Chen, J.; Tao, W.; Wang, K.; Zheng, C.; Liu, W.; Li, X.; Ou, X.; Zhang, X. Highly efficient thermally activated delayed fluorescence emitters based on novel Indolo[2,3-b] acridine electron-donor. Org. Electron. 2018, 57, 327–334. [Google Scholar] [CrossRef]
- Ren, C.; Zhao, F.; Kang, D.; Yang, G.; Han, X.; Tong, X.; Feng, Y.; Ren, G. Linkages of C:N:P stoichiometry and bacterial community in soil following afforestation of former farmland. Forest Ecol. Manag. 2016, 376, 59–66. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2013, 42, D643–D648. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Larsson, K.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Jing, Y.; Zhang, Y.; Han, I.; Wang, P.; Mei, Q.; Huang, Y. Effects of different straw biochars on soil organic carbon, nitrogen, available phosphorus, and enzyme activity in paddy soil. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Fu, S.; Zhang, X.; Zhao, K.; Chen, H.Y.H. Intercropping improves soil nutrient availability, soil enzyme activity and tea quantity and quality. Appl. Soil Ecol. 2017, 119, 171–178. [Google Scholar] [CrossRef]
- Curtright, A.J.; Tiemann, L.K. Intercropping increases soil extracellular enzyme activity: A meta-analysis. Agr. Ecosyst. Environ. 2021, 319, 107489. [Google Scholar] [CrossRef]
- Mouradi, M.; Farissi, M.; Makoudi, B.; Bouizgaren, A.; Ghoulam, C. Effect of faba bean (Vicia faba L.)–rhizobia symbiosis on barley's growth, phosphorus uptake and acid phosphatase activity in the intercropping system. Ann. Agrar. Sci. 2018, 16, 297–303. [Google Scholar] [CrossRef]
- Chen, X.; Song, B.; Yao, Y.; Wu, H.; Hu, J.; Zhao, L. Aromatic plants play an important role in promoting soil biological activity related to nitrogen cycling in an orchard ecosystem. Sci. Total Environ. 2014, 472, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cao, F. Integrated evaluation of soil fertility in Ginkgo (Ginkgo biloba L.) agroforestry systems in Jiangsu, China. Agroforest Syst. 2011, 83, 89–100. [Google Scholar] [CrossRef]
- Fan, L.; Tarin, M.W.K.; Zhang, Y.; Han, Y.; Rong, J.; Cai, X.; Chen, L.; Shi, C.; Zheng, Y. Patterns of soil microorganisms and enzymatic activities of various forest types in coastal sandy land. Glob. Ecol. Conserv. 2021, 28, e1625. [Google Scholar] [CrossRef]
- Menezes, K.M.S.; Silva, D.K.A.; Gouveia, G.V.; Da Costa, M.M.; Queiroz, M.A.A.; Yano-Melo, A.M. Shading and intercropping with buffelgrass pasture affect soil biological properties in the Brazilian semi-arid region. Catena 2019, 175, 236–250. [Google Scholar] [CrossRef]
- Li, N.; Gao, D.; Zhou, X.; Chen, S.; Li, C.; Wu, F. Intercropping with Potato-Onion Enhanced the Soil Microbial Diversity of Tomato. Microorganisms 2020, 8, 834. [Google Scholar] [CrossRef]
- Li, S.; Wu, F. Diversity and Co-occurrence Patterns of Soil Bacterial and Fungal Communities in Seven Intercropping Systems. Front. Microbiol. 2018, 9, 1521. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Khan, M.U.; Lin, X.; Lin, Z.; Lin, S.; Lin, W. Evaluation of maize/peanut intercropping effects on microbial assembly, root exudates and peanut nitrogen uptake. Plant Physiol. Bioch. 2022, 171, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, X.; Zhang, J.; Wei, H.; Li, M.; Lan, N.; Luo, H. Intercropping perennial aquatic plants with rice improved paddy field soil microbial biomass, biomass carbon and biomass nitrogen to facilitate soil sustainability. Soil Tillage Res. 2021, 208, 104908. [Google Scholar] [CrossRef]
- Singh, S.R.; Yadav, P.; Singh, D.; Shukla, S.K.; Tripathi, M.K.; Bahadur, L.; Mishra, A.; Kumar, S. Intercropping in Sugarcane Improves Functional Diversity, Soil Quality and Crop Productivity. Sugar Tech 2021, 1–17. [Google Scholar] [CrossRef]
- Cheng, H.; Yuan, M.; Tang, L.; Shen, Y.; Yu, Q.; Li, S. Integrated microbiology and metabolomics analysis reveal responses of soil microorganisms and metabolic functions to phosphorus fertilizer on semiarid farm. Sci. Total Environ. 2022, 817, 152878. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Wang, M.; Si, T.; Wang, Y.; Zhao, H.; Zhang, X.; Yu, X.; Wan, S.; Zou, X. Maize-peanut intercropping led to an optimization of soil from the perspective of soil microorganism. Arch. Acker-Pflanzenbau Bodenkd. 2021, 67, 1986–1999. [Google Scholar] [CrossRef]
- Mouhamadou, B.; Puissant, J.; Personeni, E.; Desclos-Theveniau, M.; Kastl, E.M.; Schloter, M.; Zinger, L.; Roy, J.; Geremia, R.A.; Lavorel, S. Effects of two grass species on the composition of soil fungal communities. Biol. Fert. Soils 2013, 49, 1131–1139. [Google Scholar] [CrossRef]
- Yang, J.; Duan, Y.; Liu, X.; Sun, M.; Wang, Y.; Liu, M.; Zhu, Z.; Shen, Z.; Gao, W.; Wang, B.; et al. Reduction of banana fusarium wilt associated with soil microbiome reconstruction through green manure intercropping. Agric. Ecosyst. Environ. 2022, 337, 108065. [Google Scholar] [CrossRef]
- Yin, Y.; Yang, C.; Tang, J.; Gu, J.; Li, H.; Duan, M.; Wang, X.; Chen, R. Bamboo charcoal enhances cellulase and urease activities during chicken manure composting: Roles of the bacterial community and metabolic functions. J. Environ. Sci.-China 2021, 108, 84–95. [Google Scholar] [CrossRef]
- Fan, F.; Yin, C.; Tang, Y.; Li, Z.; Song, A.; Wakelin, S.A.; Zou, J.; Liang, Y. Probing potential microbial coupling of carbon and nitrogen cycling during decomposition of maize residue by 13C-DNA-SIP. Soil Biol. Biochem. 2014, 70, 12–21. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolo, P.; Kihara, J.; Mucheru-Muna, M.; Njeru, E.M.; Kinyua, M.; Sommer, R. Application of residue, inorganic fertilizer and lime affect phosphorus solubilizing microorganisms and microbial biomass under different tillage and cropping systems in a Ferralsol. Geoderma 2021, 390, 114962. [Google Scholar] [CrossRef]
- Yang, Y.; Tong, Y.; Liang, L.; Li, H.; Han, W. Dynamics of soil bacteria and fungi communities of dry land for 8 years with soil conservation management. J. Environ. Manag. 2021, 299, 113544. [Google Scholar] [CrossRef] [PubMed]
- Janvier, C.; Villeneuve, F.O.; Alabouvette, C.; Edel-Hermann, V.; Mateille, T.; Steinberg, C. Soil health through soil disease suppression: Which strategy from descriptors to indicators? Soil Biol. Biochem. 2007, 39, 1–23. [Google Scholar] [CrossRef]
- Yu, L.; Luo, S.; Gou, Y.; Xu, X.; Wang, J. Structure of rhizospheric microbial community and N cycling functional gene shifts with reduced N input in sugarcane-soybean intercropping in South China. Agric. Ecosyst. Environ. 2021, 314, 107413. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, G.; Wu, Z.; Wen, X.; Zhong, H.; Zhong, Z.; Bian, F.; Gai, X. Agroforestry alters the rhizosphere soil bacterial and fungal communities of moso bamboo plantations in subtropical China. Appl. Soil Ecol. 2019, 143, 192–200. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, Y.; Feng, K.; Li, S.; Wang, S.; Jin, D.; Zhang, Y.; Chen, H.; Yin, H.; Xu, M.; et al. The divergence between fungal and bacterial communities in seasonal and spatial variations of wastewater treatment plants. Sci. Total Environ. 2018, 628-629, 969–978. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, X.; Huang, X.; Huang, C.; Wang, H.; Yin, H.; Shao, Y.; Li, P. Linking microbial community composition to farming pattern in selenium-enriched region: Potential role of microorganisms on Se geochemistry. J. Environ. Sci.-China 2022, 112, 269–279. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, L.; Wang, J.; Zhang, Y.; Xiao, C. Effects of warming on the bacterial community and its function in a temperate steppe. Sci. Total Environ. 2021, 792, 148409. [Google Scholar] [CrossRef]
- Song, Z.; Kennedy, P.G.; Feng, J.L.; Schilling, J.S. Fungal endophytes as priority colonizers initiating wood decomposition. Funct. Ecol. 2016, 31, 407–418. [Google Scholar] [CrossRef]


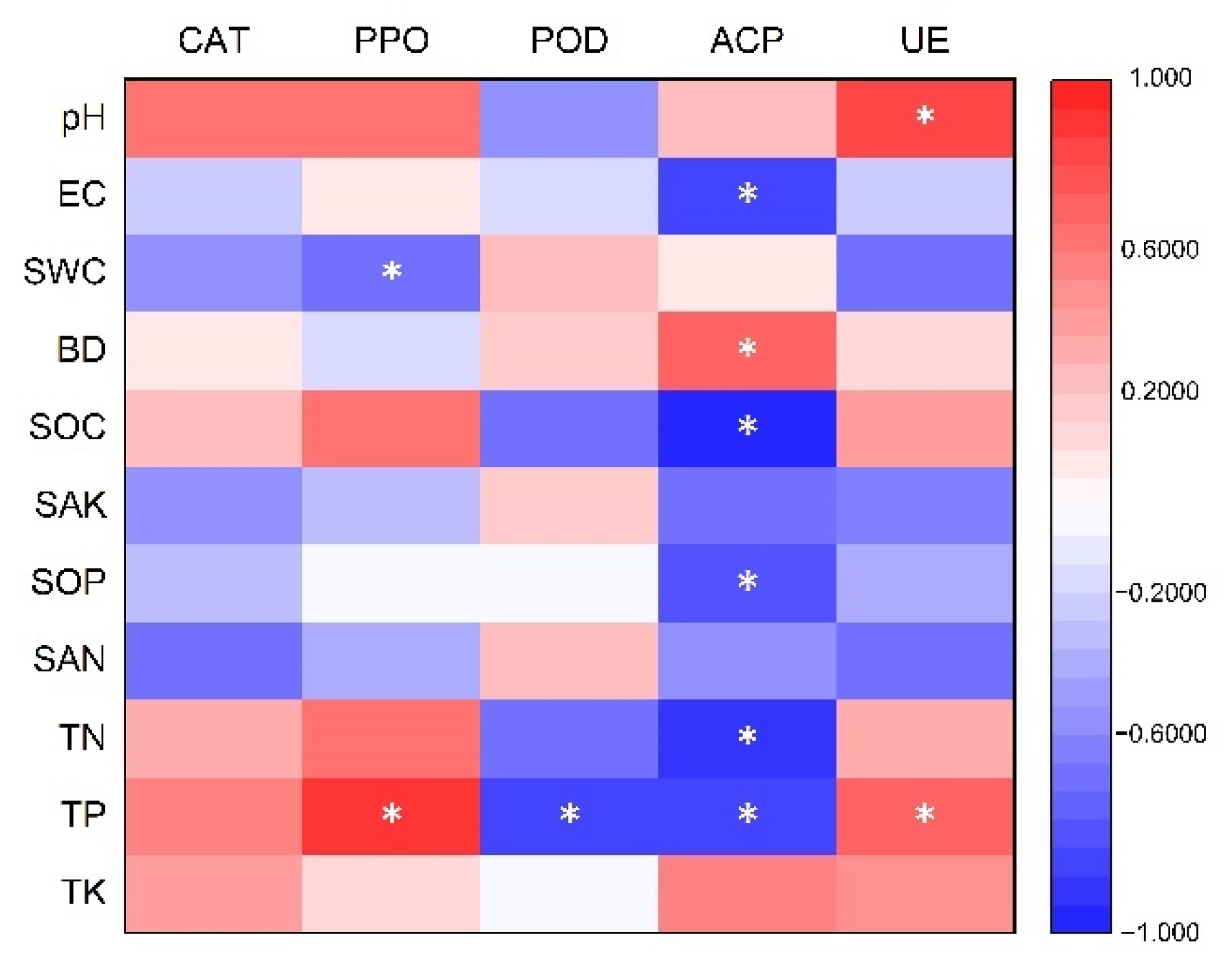
| Soil Properties | Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| Shannon | Simpson | Ace | Chao | Shannon | Simpson | Ace | Chao | |
| pH | −0.19 | 0.17 | 0.12 | 0.10 | −0.53 | 0.30 | −0.45 | −0.46 |
| EC | −0.31 | 0.28 | −0.64 | −0.63 | 0.67 * | −0.60 | −0.26 | −0.24 |
| SWC | 0.05 | −0.03 | −0.02 | 0.00 | 0.16 | −0.02 | 0.48 | 0.49 |
| BD | 0.28 | −0.16 | 0.60 | 0.59 | −0.58 | 0.59 | 0.26 | 0.25 |
| SOC | −0.61 | 0.58 | −0.79 * | −0.80 ** | 0.56 | −0.65 | −0.75 * | −0.74 * |
| SAK | −0.16 | 0.14 | −0.48 | −0.46 | 0.68 * | −0.53 | 0.11 | 0.12 |
| SOP | −0.31 | 0.27 | −0.64 | −0.63 | 0.73 * | −0.65 | −0.15 | −0.14 |
| SAN | −0.09 | 0.06 | −0.39 | −0.37 | 0.54 | −0.37 | 0.21 | 0.22 |
| TN | −0.42 | 0.38 | −0.64 | −0.66 | 0.59 | −0.66 | −0.68 * | −0.67 * |
| TP | −0.63 | 0.57 | −0.71 * | −0.73 * | 0.35 | −0.53 | −0.91 *** | −0.91 *** |
| TK | 0.27 | −0.25 | 0.62 | 0.61 | −0.40 | 0.35 | 0.36 | 0.34 |
| CAT | −0.11 | 0.09 | −0.05 | −0.09 | −0.16 | −0.02 | −0.69 * | −0.70 * |
| PPO | −0.50 | 0.45 | −0.47 | −0.50 | 0.04 | −0.27 | −0.91 *** | −0.91 *** |
| POD | 0.50 | −0.50 | 0.52 | 0.54 | 0.07 | 0.13 | 0.88 ** | 0.88 ** |
| ACP | 0.64 | −0.59 | 0.83 ** | 0.84 ** | −0.76 * | 0.82 ** | 0.64 | 0.63 |
| UE | −0.48 | 0.45 | −0.30 | −0.33 | −0.22 | −0.03 | −0.84 ** | −0.84 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Y.; Zhang, A.; Qin, X.; Yu, H.; Ji, X.; He, S.; Zong, Y.; Wang, J.; Tang, J. Effects of Intercropping Pandanus amaryllifolius on Soil Properties and Microbial Community Composition in Areca Catechu Plantations. Forests 2022, 13, 1814. https://doi.org/10.3390/f13111814
Zhong Y, Zhang A, Qin X, Yu H, Ji X, He S, Zong Y, Wang J, Tang J. Effects of Intercropping Pandanus amaryllifolius on Soil Properties and Microbial Community Composition in Areca Catechu Plantations. Forests. 2022; 13(11):1814. https://doi.org/10.3390/f13111814
Chicago/Turabian StyleZhong, Yiming, Ang Zhang, Xiaowei Qin, Huan Yu, Xunzhi Ji, Shuzhen He, Ying Zong, Jue Wang, and Jinxuan Tang. 2022. "Effects of Intercropping Pandanus amaryllifolius on Soil Properties and Microbial Community Composition in Areca Catechu Plantations" Forests 13, no. 11: 1814. https://doi.org/10.3390/f13111814




