Comparison of Data Grouping Strategies on Prediction Accuracy of Tree-Stem Taper for Six Common Species in the Southeastern US
Abstract
:1. Introduction
2. Materials and Methodology
2.1. Data
2.2. Taper Model
2.2.1. Variable-Exponent Model
2.2.2. Segmented Polynomial Regression Model
2.3. Model Fitting and Evaluation
- A random sample of 100 trees was selected from the original dataset for a given species.
- (a)
- Population-level case (fitting a single stem taper model for all data):For a given species, 100 sample trees selected in step 1 were randomly split into fitting and validation datasets based on step 3. Then, the randomly-split trees were merged into a fitting and validation dataset, respectively. In this case, fitting and validation datasets included all species, and each species contributed equal number of trees.
- (b)
- Data grouping cases (fitting stem taper with grouped data):Trees drawn from step 1 were grouped based on taxonomic rank, tree form and size, which were detailed in Section 2.3.1 and Section 2.3.2.
- Fitting and validation data were created with 10/90, 20/80, 30/70, 40/60, 50/50, 60/40, 70/30, 80/20 and 90/10 splits. Trees used in fitting and validation were randomly selected. Model parameters were estimated with the Levenberg–Marquardt (LM) non-linear least squares algorithm that is implemented in the nlsLM function in R [17]. The LM algorithm is a compromise between the gradient-descent and Gauss–Newton approaches, which leads to more stable parameter estimates [18]. The initial values for parameter estimation were obtained from Yang and Burkhart [9]. The model evaluation statistics are given in Section 2.3.3.
- Steps 1–3 were repeated 500 times.
2.3.1. Grouping Data Based on Taxonomic Rank
- Trees grouped by species (fitting stem taper at species level):All trees selected in step 1 were used in model fitting and evaluation where fitting and validation datasets included only a single species. For a given species, 100 sample trees selected in step 1 were randomly split into fitting and validation datasets based on step 3.
- Trees grouped by species group:Six species were divided into three species groups: pine (shortleaf pine and Virginia pine), oak (white oak and southern red oak) and other hardwoods (yellow poplar and hickory spp.). Species in the pine or oak groups belong to the same genus. Although yellow popular and hickory spp. were in different genera, the classifying strategy is commonly implemented in practice when species data are not available [4]. For a given group, the fitting/validation datasets were composed of equal proportion of sample trees from each species. For example, under the 90/10 split, each species in a group contributed 90 trees for fitting and 10 trees for validation.
- Trees grouped by division group (gymnosperm vs. angiosperm):Six species were divided into softwood and hardwood groups. The softwood group included short-leaf pine and Virginia pine, whereas the other group contained white oak, southern red oak, yellow poplar and hickory spp. For a given group, the fitting/validation datasets were composed of equal proportion of sample trees from each species. Each species in a group has 100 trees randomly selected for fitting and validation.
2.3.2. Grouping Data Based on Tree Form and Size
- Six HD ratio or DBH groups: Trees were divided into six groups based on H–D ratios or DBH. Each group included 100 trees (i.e., 100 trees/group = 600 trees/6 groups).
- Three H–D ratio or DBH groups: Trees were divided into the smallest, middle and largest one-thirds based on H–D ratios or DBH to generate three H–D ratio or DBH groups. Each group included 200 trees (i.e., 200 trees/group = 600 trees/3 groups).
- Two H–D ratio or DBH groups: Trees were divided into the smallest and largest 50% based on H–D ratios or DBH to generate two H–D ratio or DBH groups. Each group included 300 trees (i.e., 300 trees/group = 600 trees/2 groups).
2.3.3. Statistics for Model Evaluation
3. Results
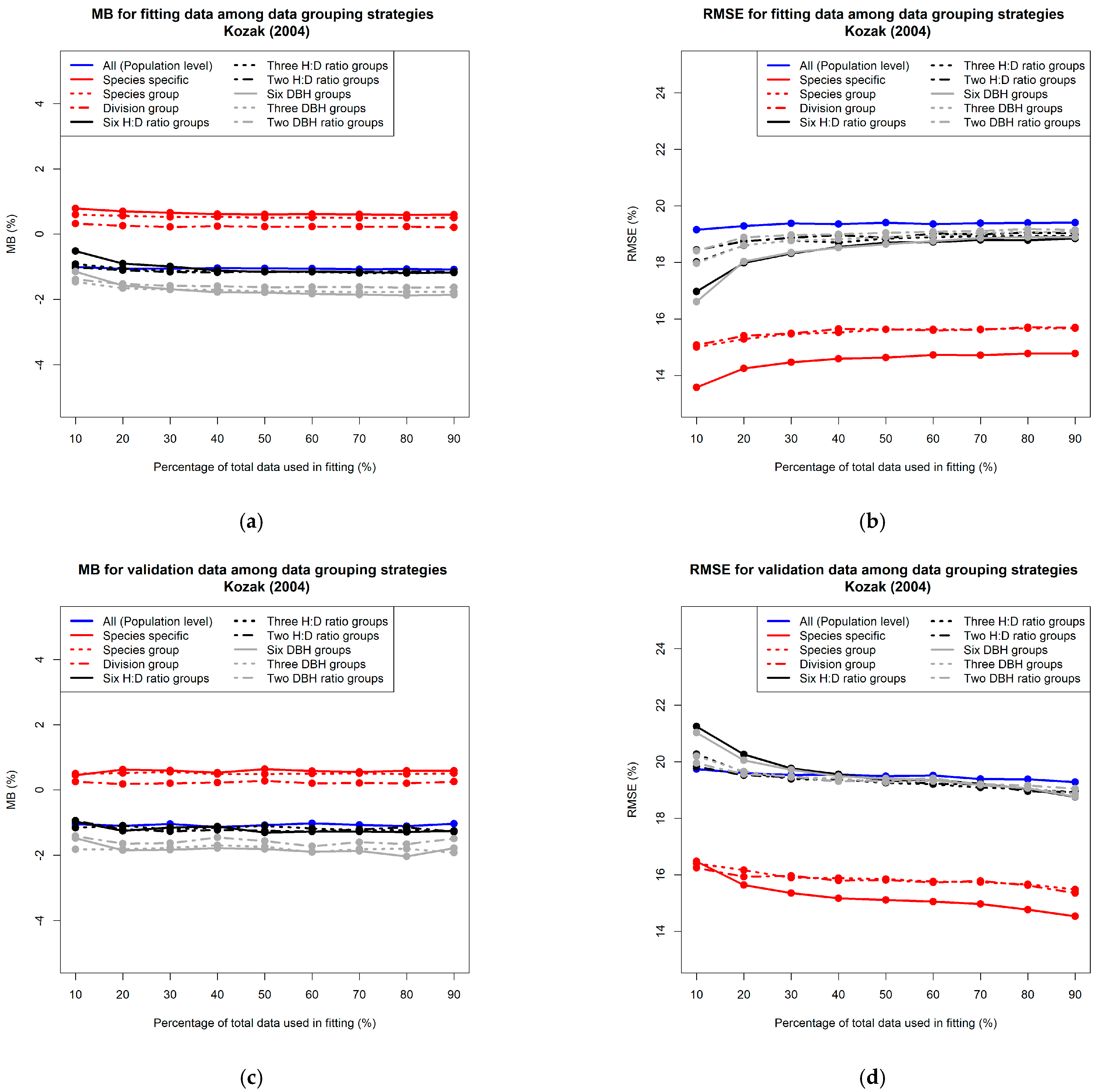
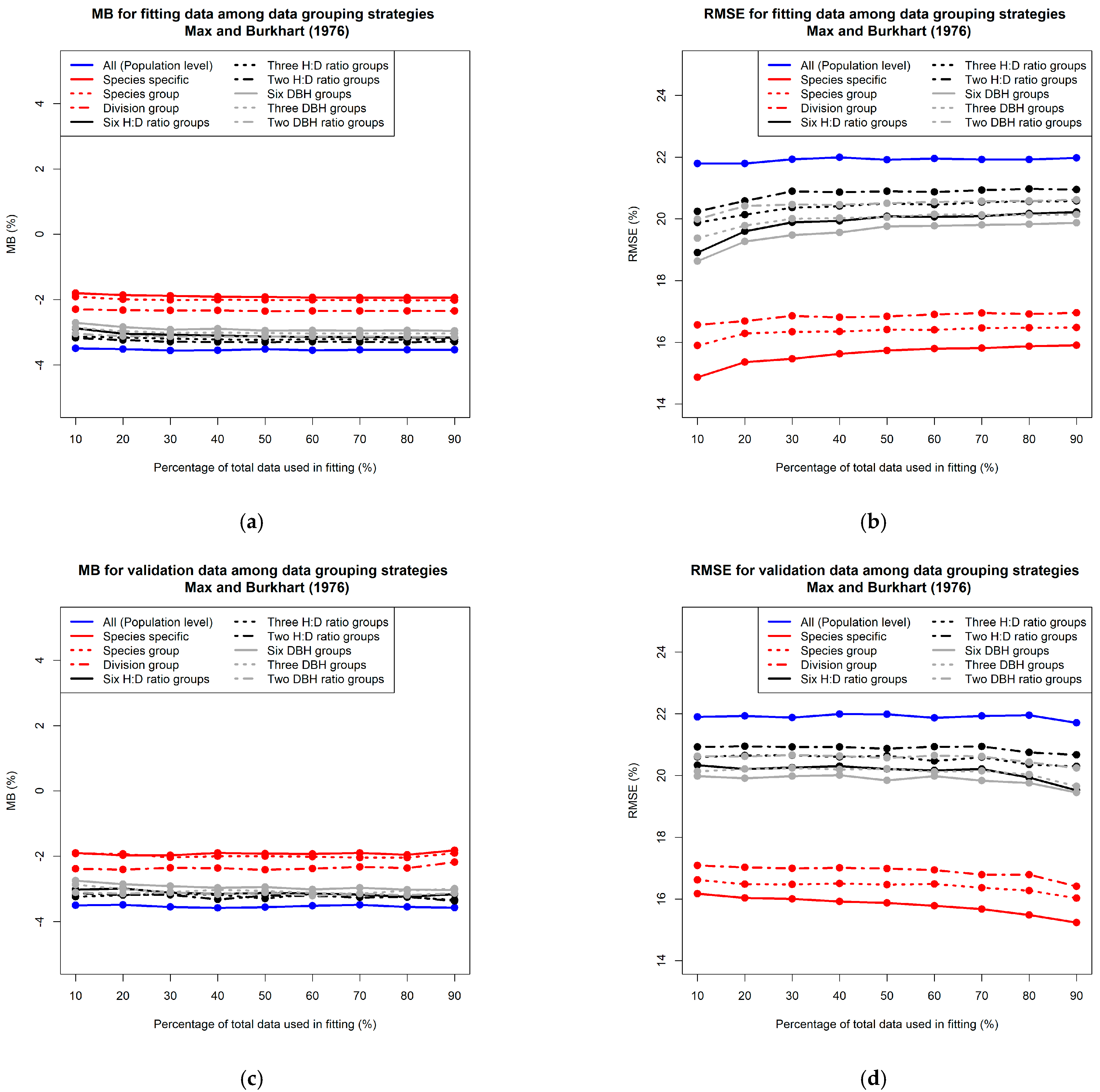
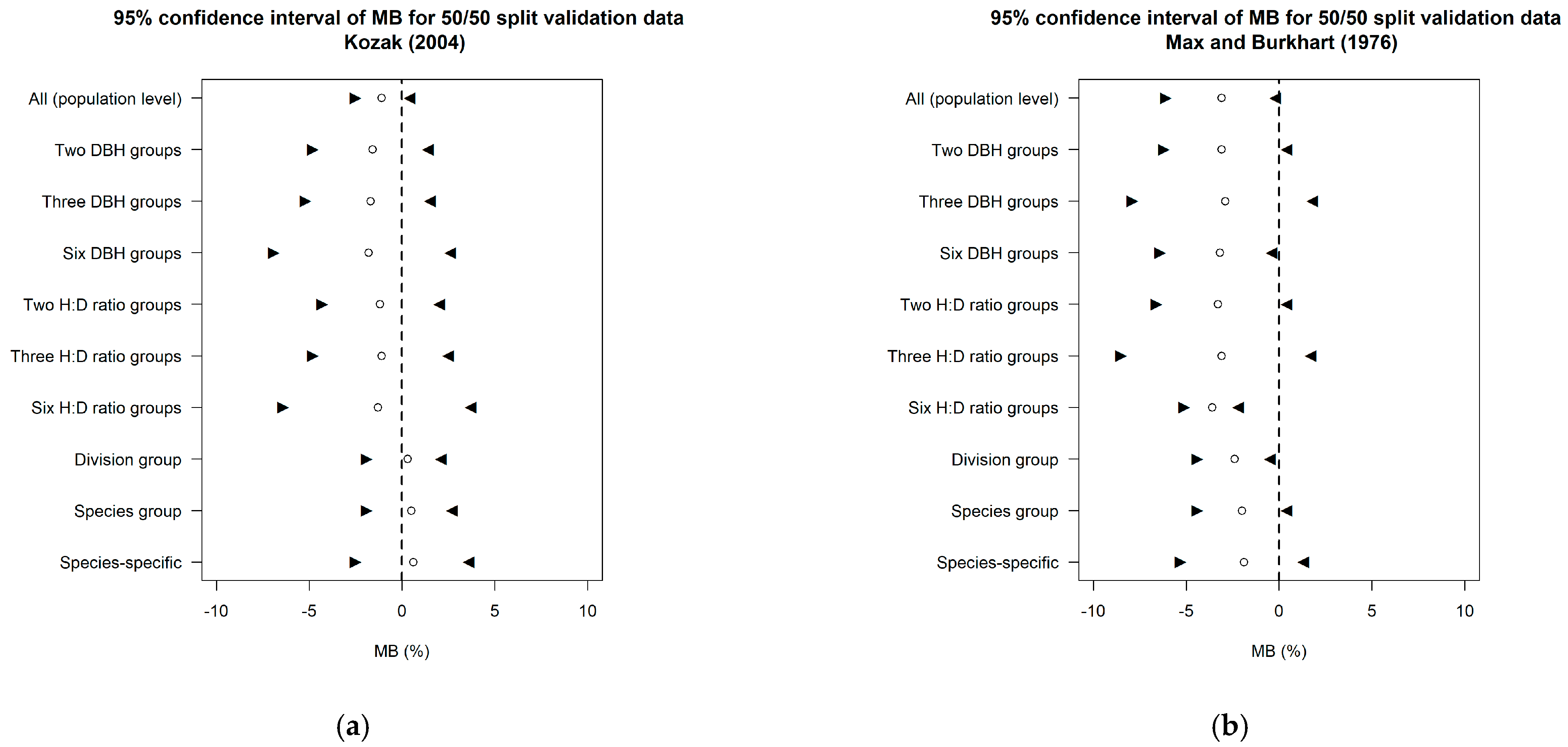
3.1. Comparison of Prediction Accuracy among Different Data Grouping Strategies
3.1.1. Grouping Data Based on Taxonomic Rank
3.1.2. Grouping Data Based on Tree Form and Size
3.2. Effect of Sample Size on Prediction Accuracy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burkhart, H.E.; Tomé, M. Modeling Forest Trees and Stands; Springer: Dordrecht, The Netherland, 2012; p. 458. [Google Scholar]
- Özçelik, R.; Brooks, J.R.; Jiang, L. Modeling Stem Profile of Lebanon cedar, Brutian pine, and Cilicica fir in Southern Turkey Using Nonlinear Mixed-effects Models. Eur. J. For. Res. 2011, 130, 613–621. [Google Scholar] [CrossRef]
- Sharma, M.; Burkhart, H.E.; Amateis, R.L. Scaling Taper Relationships from Miniature-Scale to Operational-Scale Stands of Loblolly Pine. For. Sci. 2007, 53, 611–617. [Google Scholar]
- Clark, A.; Souter, R.A.; Schlaegel, B.E. Stem Profile for Southern Equations for Southern Tree Species; Research Paper SE-282; U.S. Department of Agriculture, Forest Service, Southeastern Forest Experiment Station: Asheville, NC, USA, 1991; 117p. [CrossRef]
- Olden, J.D. A Species-Specific Approach to Modeling Biological Communities and Its Potential for Conservation. Conserv. Biol. 2003, 17, 854–863. [Google Scholar] [CrossRef] [Green Version]
- Ferrier, S.; Guisan, A. Spatial modelling of biodiversity at the community level. J. Appl. Ecol. 2006, 43, 393–404. [Google Scholar] [CrossRef]
- Kitikidou, K.; Chatzilazarou, G. Estimating the sample size for fitting taper equations. J. For. Sci. 2008, 54, 176–182. [Google Scholar] [CrossRef] [Green Version]
- MacFarlane, D.W.; Weiskittel, A.R. A new method for capturing stem taper variation for trees of diverse morphological types. Can. J. For. Res. 2016, 46, 804–815. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.I.; Burkhart, H.E. Robustness of parametric and nonparametric fitting procedures of tree-stem taper with alternative definitions for validation data. J. For. 2020, 118, 576–583. [Google Scholar] [CrossRef]
- McTague, J.P.; Weiskittel, A. Evolution, history, and use of stem taper equations: A review of their development, application, and implementation. Can. J. For. Res. 2021, 51, 210–235. [Google Scholar] [CrossRef]
- Max, T.; Burkhart, H. Segmented Polynomial Regression Applied to Taper Equations. For. Sci. 1976, 22, 283–289. [Google Scholar] [CrossRef]
- Kozak, A. My last words on taper equations. For. Chron. 2004, 80, 507–515. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Weiskittel, A.R. Comparison of model forms for estimating stem taper and volume in the primary conifer species of the North American Acadian Region. Ann. For. Sci. 2010, 67, 302. [Google Scholar] [CrossRef] [Green Version]
- Burns, R.M.; Honkala, B.H. Silvics Manual Volume 2: Hardwoods. In Agriculture Handbook 654; United States Department of Agriculture (USDA), Forest Service: Washington, DC, USA, 1990; 877p. [Google Scholar]
- Radtke, P.J.; Walker, D.M.; Weiskittel, A.R.; Frank, J.; Coulston, J.W.; Westfall, J.A. Legacy Tree Data: A National Database of Detailed Tree Measurements for Volume, Weight, and Physical Properties; General Technical Report PNW-GTR-931; U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station: Portland, OR, USA, 2015; pp. 25–30.
- USDA Forest Service. Field Guides for Standard (Phase 2) Measurements; Forest Inventory and Analysis National Program: Washington, DC, USA, 2020; p. 449. [Google Scholar]
- Elzhov, V.; Mullen, K.M.; Spiess, A.N.; Bolker, B. Package ’minpack.lm’—R Interface to the Levenberg-Marquardt Nonlinear Least-Squares Algorithm Found in MINPACK, Plus Support for Bounds. R Package Version 1.2-1. 2016. Available online: https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.192.5978&rep=rep1&type=pdf (accessed on 18 October 2021).
- Bates, D.M.; Watts, D.G. Nonlinear Regression Analysis and Its Applications; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1988; Volume 365. [Google Scholar]
- Hein, S.; Mäkinen, H.; Yue, C.; Kohnle, U. Modelling branch characteristics of Norway spruce from wide spacings in Germany. For. Ecol. Manag. 2007, 242, 155–164. [Google Scholar] [CrossRef]
- Šebeň, V.; Bošel’A, M.; Konôpka, B.; Pajtík, J. Indices of tree competition in dense spruces stand originated from natural regeneration. For. J. 2013, 59, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Phillips, P.D.; Yasman, I.; Brash, T.E.; Van Gardingen, P.R. Grouping tree species for analysis of forest data in Kalimantan (Indonesian Borneo). For. Ecol. Manag. 2002, 157, 205–216. [Google Scholar] [CrossRef]
- Gourlet-Fleury, S.; Blanc, L.; Picard, N.; Sist, P.; Dick, J.; Nasi, R.; Swaine, M.D.; Forni, E. Grouping species for predicting mixed tropical forest dynamics: Looking for a strategy. Ann. For. Sci. 2005, 62, 785–796. [Google Scholar] [CrossRef] [Green Version]
- Sabatia, C.O.; Burkhart, H.E. On the use of upper stem diameters to localize a segmented taper equation to new trees. For. Sci. 2015, 61, 411–423. [Google Scholar] [CrossRef]
- Lam, T.Y.; Kershaw, J.A.; Hajar, Z.S.N.; Rahman, K.A.; Weiskittel, A.R.; Potts, M.D. Evaluating and modelling genus and species variation in height-to-diameter relationships for Tropical Hill Forests in Peninsular Malaysia. Forestry 2017, 90, 268–278. [Google Scholar] [CrossRef] [Green Version]

| Species | Ntree | Nobs/tree | DBH (cm) | Ht (m) |
|---|---|---|---|---|
| shortleaf pine (Pinus echinata Mill.) | 1347 | 24 (4) | 35.2 (10.5) | 21.8 (3.8) |
| Virginia pine (Pinus virginiana Mill.) | 345 | 22 (3) | 29.2 (7.5) | 20.6 (3.4) |
| white oak (Quercus alba L.) | 717 | 25 (4) | 36.9 (10.0) | 23.7 (3.5) |
| southern red oak (Quercus falcata Michx.) | 292 | 24 (3) | 37.1 (8.6) | 22.4 (2.9) |
| yellow poplar (Liriodendron tulipifera L.) | 399 | 28 (4) | 40.0 (11.7) | 28.5 (4.8) |
| hickory spp. (Carya spp.) | 578 | 25 (4) | 35.4 (10.2) | 24.0 (3.9) |
| Mean Bias (%) | RMSE (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | Species | Grouping | 10/90 | 50/50 | 90/10 | 10/90 | 50/50 | 90/10 |
| Kozak (2004) | shortleaf pine (Pinus echinata Mill.) | Species-specific | −1.3 | −0.8 | −0.8 | 14.3 | 12.7 | 11.8 |
| Species group | 3.2 | 2.8 | 2.9 | 13.4 | 12.5 | 11.8 | ||
| Division group | 3.0 | 2.9 | 2.8 | 13.4 | 12.5 | 11.7 | ||
| Population level | 15.0 | 15.1 | 15.0 | 23.5 | 23.4 | 23.3 | ||
| Virginia pine (Pinus virginiana Mill.) | Species-specific | −1.0 | −0.6 | −0.9 | 14.3 | 13.1 | 12.7 | |
| Species group | −4.9 | −5.2 | −5.1 | 17.6 | 17.3 | 17.0 | ||
| Division group | −5.3 | −5.2 | −5.0 | 17.8 | 17.5 | 16.7 | ||
| Population level | 5.7 | 5.6 | 5.9 | 15.8 | 15.3 | 15.1 | ||
| yellow poplar (Liriodendron tulipifera L.) | Species-specific | 0.9 | 1.0 | 0.9 | 15.2 | 13.9 | 13.2 | |
| Species group | 1.6 | 1.6 | 1.6 | 15.5 | 14.7 | 13.9 | ||
| Division group | 2.1 | 2.1 | 2.3 | 16.1 | 15.6 | 14.9 | ||
| Population level | −2.9 | −3.1 | −2.7 | 15.7 | 15.3 | 14.0 | ||
| white oak (Quercus alba L.) | Species-specific | 1.6 | 1.7 | 1.8 | 17.7 | 16.3 | 15.7 | |
| Species group | 1.5 | 1.9 | 2.0 | 17.3 | 16.6 | 16.3 | ||
| Division group | 1.9 | 1.9 | 1.9 | 16.8 | 16.2 | 15.8 | ||
| Population level | −6.5 | −6.5 | −6.3 | 19.7 | 19.1 | 18.5 | ||
| southern red oak (Quercus falcata Michx.) | Species-specific | 1.5 | 1.7 | 1.6 | 18.2 | 17.0 | 16.7 | |
| Species group | 1.3 | 1.3 | 1.4 | 17.0 | 16.7 | 16.6 | ||
| Division group | 1.2 | 1.0 | 1.0 | 16.7 | 16.4 | 15.8 | ||
| Population level | −8.7 | −8.5 | −8.5 | 21.4 | 21.1 | 20.6 | ||
| hickory spp. (Carya spp.) | Species-specific | 1.0 | 0.9 | 1.0 | 19.2 | 17.5 | 17.2 | |
| Species group | 0.3 | 0.0 | 0.2 | 17.2 | 16.7 | 16.2 | ||
| Division group | 1.0 | 0.9 | 1.0 | 17.5 | 17.2 | 16.6 | ||
| Population level | −7.3 | −7.3 | −7.2 | 21.0 | 20.7 | 20.2 | ||
| Max and Burkhart (1976) | shortleaf pine (Pinus echinata Mill.) | Species-specific | −2.9 | −2.7 | −2.5 | 15.6 | 15.2 | 14.3 |
| Species group | 0.7 | 0.8 | 1.0 | 13.9 | 13.7 | 13.1 | ||
| Division group | 0.8 | 0.7 | 1.2 | 14.0 | 13.6 | 12.7 | ||
| Population level | 13.4 | 13.3 | 13.3 | 20.1 | 19.9 | 19.6 | ||
| Virginia pine (Pinus virginiana Mill.) | Species-specific | −3.6 | −3.5 | −3.6 | 18.1 | 18.2 | 17.7 | |
| Species group | −7.7 | −7.7 | −7.1 | 21.8 | 21.7 | 21.0 | ||
| Division group | −7.6 | −7.7 | −7.2 | 21.9 | 21.7 | 20.7 | ||
| Population level | 5.7 | 5.6 | 5.7 | 15.2 | 15.1 | 14.8 | ||
| yellow poplar (Liriodendron tulipifera L.) | Species-specific | −2.1 | −2.1 | −1.9 | 16.0 | 15.9 | 14.9 | |
| Species group | −0.4 | −0.6 | −0.4 | 14.9 | 14.7 | 13.7 | ||
| Division group | 0.4 | 0.5 | 0.7 | 14.6 | 14.4 | 13.5 | ||
| Population level | −7.4 | −7.5 | −7.6 | 19.9 | 19.8 | 19.2 | ||
| white oak (Quercus alba L.) | Species-specific | −0.9 | −1.0 | −0.9 | 15.4 | 14.9 | 14.3 | |
| Species group | −0.2 | −0.3 | −0.1 | 15.1 | 14.7 | 14.0 | ||
| Division group | −1.7 | −1.5 | −1.4 | 15.8 | 15.3 | 14.6 | ||
| Population level | −9.3 | −9.3 | −8.9 | 23.6 | 23.6 | 22.5 | ||
| southern red oak (Quercus falcata Michx.) | Species-specific | −0.9 | −1.1 | −1.0 | 15.4 | 15.0 | 14.6 | |
| Species group | −1.7 | −1.7 | −1.8 | 15.4 | 15.2 | 14.5 | ||
| Division group | −3.2 | −3.2 | −3.1 | 16.4 | 16.3 | 15.6 | ||
| Population level | −11.1 | −11.2 | −11.1 | 25.5 | 25.6 | 25.1 | ||
| hickory spp. (Carya spp.) | Species-specific | −1.1 | −1.2 | −1.0 | 16.5 | 16.2 | 15.7 | |
| Species group | −2.8 | −3.0 | −3.0 | 17.7 | 17.7 | 17.1 | ||
| Division group | −2.1 | −2.1 | −2.0 | 16.8 | 16.7 | 15.8 | ||
| Population level | −9.7 | −9.9 | −9.6 | 24.5 | 24.8 | 23.8 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.-I.; Green, P.C. Comparison of Data Grouping Strategies on Prediction Accuracy of Tree-Stem Taper for Six Common Species in the Southeastern US. Forests 2022, 13, 156. https://doi.org/10.3390/f13020156
Yang S-I, Green PC. Comparison of Data Grouping Strategies on Prediction Accuracy of Tree-Stem Taper for Six Common Species in the Southeastern US. Forests. 2022; 13(2):156. https://doi.org/10.3390/f13020156
Chicago/Turabian StyleYang, Sheng-I, and P. Corey Green. 2022. "Comparison of Data Grouping Strategies on Prediction Accuracy of Tree-Stem Taper for Six Common Species in the Southeastern US" Forests 13, no. 2: 156. https://doi.org/10.3390/f13020156






