Effects of Plant Fine Root Functional Traits and Soil Nutrients on the Diversity of Rhizosphere Microbial Communities in Tropical Cloud Forests in a Dry Season
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Samples Collection
2.3. Selection and Measurement of Fine Root Functional Traits
2.4. Determination of Soil Physical and Chemical Properties
2.5. Sequencing of Rhizosphere Microorganisms
2.6. Data Analysis
3. Results
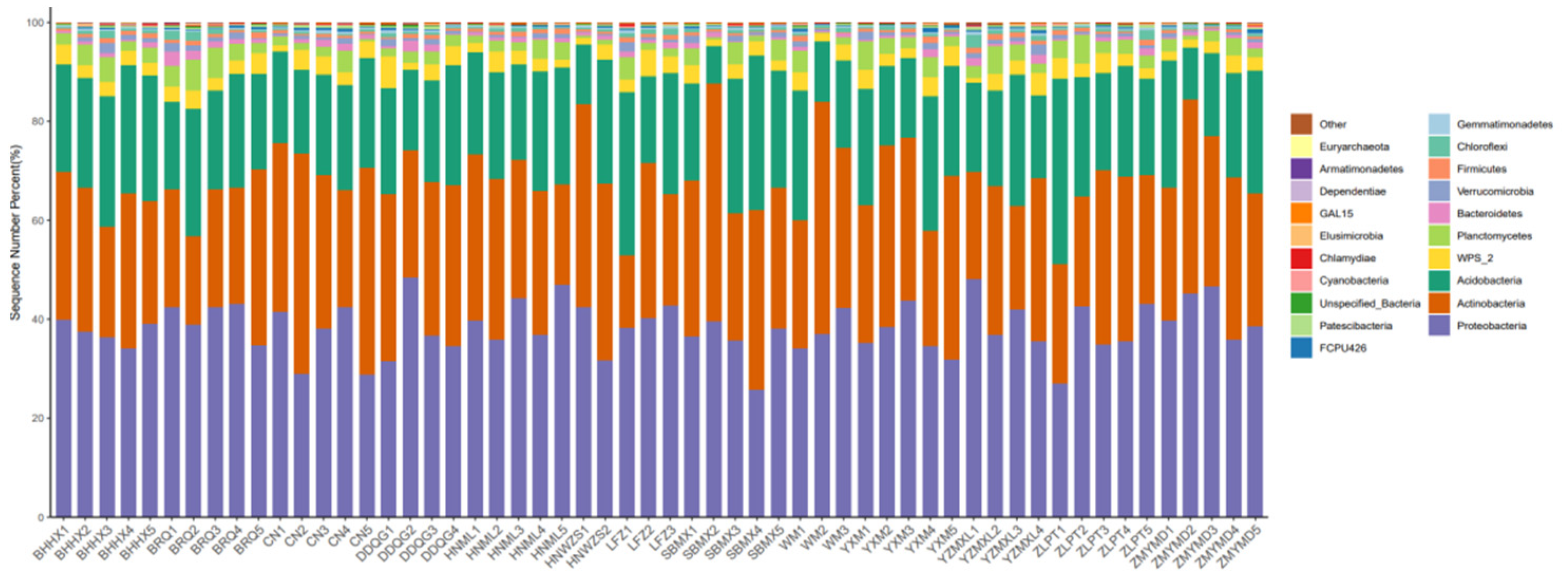
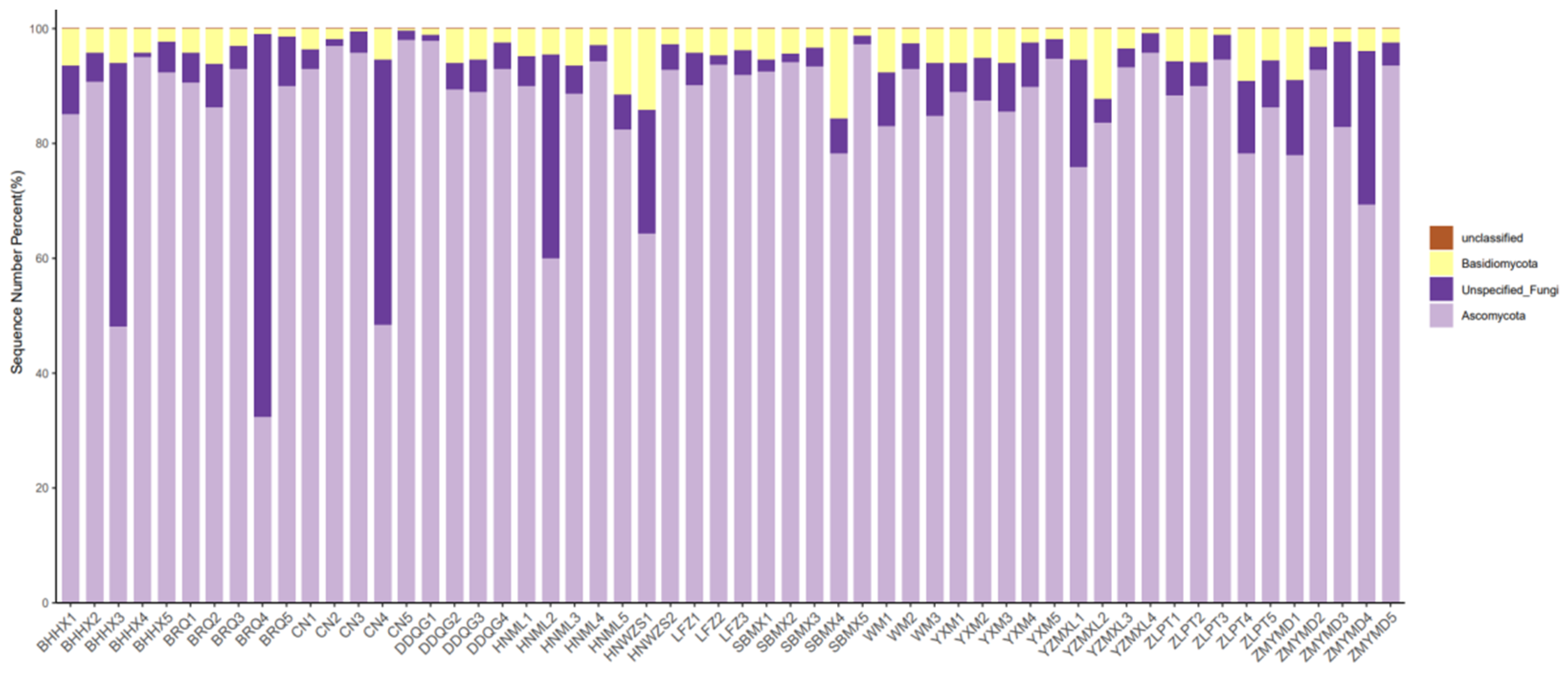
3.1. Rhizosphere Microbial Diversity
3.2. Correlation between Fine Root Functional Traits and Rhizosphere Soil
3.3. Effects of Fine Root Functional Traits and Rhizosphere Soil on Microbial Diversity
4. Discussion
4.1. Diversity of Rhizosphere Microbial Community
4.2. Impact of the Soil Environment on the Microbial Diversity of the Rhizosphere Soil Relative to the Functional Traits of Fine Roots
4.3. Soil pH Effects on Rhizosphere Soil Microbes in the Soil Environment
4.4. Effects of Soil Available Phosphorus on Rhizosphere Microorganisms
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hiltner, L. Ber neuere erfahrungen und probleme auf dem gebiete der bodenbakteriologie unter besonderer bercksichtigung der grndngung und brache. Arb. Dtsch. Landwirtsch. Ges. 1904, 98, 59–78. [Google Scholar]
- Banerjee, S.; Kaushik, S.; Tomar, R.S. Rhizobacters as Remedy of Stress Tolerance in Potato. In Antioxidants in Plant-Microbe Interaction; Bahadur Singh, H., Vaishnav, A., Eds.; Springer: Singapore, 2021; pp. 395–412. [Google Scholar]
- Zhang, P.; Cui, Z.; Guo, M.; Xi, R. Characteristics of the soil microbial community in the forestland of Camellia oleifera. PeerJ 2020, 8, e9117. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Marsh, E.L.; Ainsworth, E.A.; Leakey, A.D.; Sheflin, A.M.; Schachtman, D.P. Shifts in microbial communities in soil, rhizosphere and roots of two major crop systems under elevated CO2 and O3. Sci. Rep. 2017, 7, 15019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Wei, Z.; Hu, X.; Li, X.; Zhang, Y.; Jiang, L.; Li, J.; Guan, Z.; Cai, Y.; Liao, X. The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in Taihu Lake area, China. PLoS ONE 2017, 12, e0174411. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Hannula, S.; Morrien, E.; van der Putten, W.; de Boer, W. Rhizosphere fungi actively assimilating plant-derived carbon in a grassland soil. Fungal Ecol. 2020, 48, 100988. [Google Scholar] [CrossRef]
- Freschet, G.T.; Roumet, C. Sampling roots to capture plant and soil functions. Funct. Ecol. 2017, 31, 1506–1518. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, J.J.; Nadelhoffer, K.J.; Aber, J.D. Assessing the role of fine roots in carbon and nutrient cycling. Trends Ecol. Evol. 1993, 8, 174–178. [Google Scholar] [CrossRef]
- Sweeney, C.J.; de Vries, F.T.; van Dongen, B.E.; Bardgett, R.D. Root traits explain rhizosphere fungal community composition among temperate grassland plant species. New Phytol. 2021, 229, 1492–1507. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, C.M.; Lindahl, B.; Wardle, D.A.; Sundqvist, M.K.; Gundale, M.J.; Fanin, N.; Kardol, P. Root trait-microbial relationships across tundra plant species. New Phytol. 2021, 229, 1508–1520. [Google Scholar] [CrossRef] [PubMed]
- Merino-Martín, L.; Griffiths, R.I.; Gweon, H.S.; Furget-Bretagnon, C.; Oliver, A.; Mao, Z.; Le Bissonnais, Y.; Stokes, A. Rhizosphere bacteria are more strongly related to plant root traits than fungi in temperate montane forests: Insights from closed and open forest patches along an elevational gradient. Plant 2020, 450, 183–200. [Google Scholar] [CrossRef]
- Elhaissoufi, W.; Khourchi, S.; Ibnyasser, A.; Ghoulam, C.; Rchiad, Z.; Zeroual, Y.; Lyamlouli, K.; Bargaz, A. Phosphate solubilizing rhizobacteria could have a stronger influence on wheat root traits and aboveground physiology than rhizosphere P solubilization. Front. Plant Sci. 2020, 11, 979. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Chen, J.; Deng, J.; Zhao, F.; Han, X.; Yang, G.; Tong, X.; Feng, Y.; Shelton, S.; Ren, G. Response of microbial diversity to C:N:P stoichiometry in fine root and microbial biomass following afforestation. Biol. Fertil. Soils 2017, 53, 457–468. [Google Scholar] [CrossRef]
- Ding, X.; Liu, G.; Fu, S.; Chen, H.Y.H. Tree species composition and nutrient availability affect soil microbial diversity and composition across forest types in subtropical China. CATENA 2021, 201, 105224. [Google Scholar] [CrossRef]
- Barberán, A.; McGuire, K.L.; Wolf, J.A.; Jones, F.A.; Wright, S.J.; Turner, B.L.; Essene, A.; Hubbell, S.P.; Faircloth, B.C.; Fierer, N. Relating belowground microbial composition to the taxonomic, phylogenetic, and functional trait distributions of trees in a tropical forest. Ecol. Lett. 2015, 18, 1397–1405. [Google Scholar] [CrossRef]
- Rivest, M.; Whalen, J.K.; Rivest, D. Tree diversity is not always a strong driver of soil microbial diversity: A 7-yr-old diversity experiment with trees. Ecosphere 2019, 10, e02685. [Google Scholar] [CrossRef]
- Oh, Y.M.; Kim, M.; Lee-Cruz, L.; Lai-Hoe, A.; Go, R.; Ainuddin, N.; Rahim, R.A.; Shukor, N.; Adams, J.M. Distinctive Bacterial Communities in the Rhizoplane of Four Tropical Tree Species. Microb. Ecol. 2012, 64, 1018–1027. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Zhang, J.; Cai, Z.; Jiang, K.; Chang, Y. Deciphering the relative importance of soil and plant traits on the development of rhizosphere microbial communities. Soil Biol. Biochem. 2020, 148, 107909. [Google Scholar] [CrossRef]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, R.; Yang, X.; Tian, Q.; Wang, X.; Liao, C.; Li, C.-L.; Liu, F. Soil microbial community structure, metabolic potentials and influencing factors in a subtropical mountain forest ecosystem of China. Environ. Pollut. Bioavailab. 2020, 32, 69–78. [Google Scholar] [CrossRef]
- Glassman, I.S.; Wang, I.J.; Bruns, T.D. Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol. Ecol. 2017, 26, 6960–6973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xun, W.; Huang, T.; Zhao, J.; Ran, W.; Wang, B.; Shen, Q.; Zhang, R. Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol. Biochem. 2015, 90, 10–18. [Google Scholar] [CrossRef]
- Wan, W.; Hao, X.; Xing, Y.; Liu, S.; Zhang, X.; Li, X.; Chen, W.; Huang, Q. Spatial differences in soil microbial diversity caused by pH-driven organic phosphorus mineralization. Land Degrad. Dev. 2021, 32, 766–776. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef]
- Malik, A.A.; Puissant, J.; Buckeridge, K.M.; Goodall, T.; Jehmlich, N.; Chowdhury, S.; Gweon, H.S.; Peyton, J.M.; Mason, K.E.; van Agtmaal, M.; et al. Land use driven change in soil pH affects microbial carbon cycling processes. Nat. Commun. 2018, 9, 3591. [Google Scholar] [CrossRef]
- Loope, L.L.; Giambelluca, T.W. Vulnerability of island tropical montane cloud forests to climate change, with special reference to East Maui, Hawaii. In Potential Impacts of Climate Change on Tropical Forest Ecosystems; Markham, A., Ed.; Springer: Dordrecht, The Netherlands, 1998; pp. 363–377. [Google Scholar]
- Long, W.; Zhou, Y.; Schamp, B.S.; Zang, R.; Yang, X.; Poorter, L.; Xiao, C.; Xiong, M. Scaling relationships among functional traits are similar across individuals, species, and communities. J. Veg. Sci. 2020, 31, 571–580. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, H.; Zang, R.; Wang, X.; Long, W.; Wang, X.; Xiong, M.; John, R. The effects of soil phosphorus on aboveground biomass are mediated by functional diversity in a tropical cloud forest. Plant Soil 2020, 449, 51–63. [Google Scholar] [CrossRef]
- Long, W.; Zang, R.; Ding, Y. Air temperature and soil phosphorus availability correlate with trait differences between two types of tropical cloud forests. Flora-Morphol. Distrib. Funct. Ecol. Plants 2011, 206, 896–903. [Google Scholar] [CrossRef]
- Jiang, Y.; Zang, R.; Lu, X.; Huang, Y.; Ding, Y.; Liu, W.; Long, W.; Zhang, J.; Zhang, Z. Effects of soil and microclimatic conditions on the community-level plant functional traits across different tropical forest types. Plant Soil 2015, 390, 351–367. [Google Scholar] [CrossRef]
- Long, W.; Zang, R.; Ding, Y.; Huang, Y. Effects of Competition and Facilitation on Species Assemblage in Two Types of Tropical Cloud Forest. PLoS ONE 2013, 8, e60252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, W.; Schamp, B.S.; Zang, R.; Ding, Y.; Huang, Y.; Xiang, Y. Community assembly in a tropical cloud forest related to specific leaf area and maximum species height. J. Veg. Sci. 2015, 26, 513–523. [Google Scholar] [CrossRef]
- Barillot, C.C.D.; Sarde, C.-O.; Bert, V.; Tarnaud, E.; Cochet, N. A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann. Microbiol. 2013, 63, 471–476. [Google Scholar] [CrossRef]
- Fujita, S.; Noguchi, K.; Tange, T. Root Responses of Five Japanese Afforestation Species to Waterlogging. Forests 2020, 11, 552. [Google Scholar] [CrossRef]
- Petriţan, I.C.; von Lüpke, B.; Petriţan, A.M. Effects of root trenching of overstorey Norway spruce (Picea abies) on growth and biomass of underplanted beech (Fagus sylvatica) and Douglas fir (Pseudotsuga menziesii) saplings. Eur. J. For. Res. 2011, 130, 813–828. [Google Scholar] [CrossRef] [Green Version]
- Liu, F. Practical Manual of Agricultural Environment Monitoring; China Standards Press: Beijing, China, 2001; pp. 93–191. [Google Scholar]
- Lu, R.K. Soil Agrochemical Analysis Methods; China Agricultural Science and Technology Press: Beijing, China, 2000; p. 315. [Google Scholar]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, R.M.; Silveira ÉLd Scaquitto, D.C.; Pedrinho, E.A.N.; Val-Moraes, S.P.; Wickert, E.; Carareto-Alves, L.M.; de Macedo Lemos, E.G. Molecular characterization of bacterial populations of different soils. Braz. J. Microbiol. 2006, 37, 439–447. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Li, Y.; Ye, C.; He, X.; Zhang, S. Fate of antibiotics and antibiotic resistance genes during aerobic co-composting of food waste with sewage sludge. Sci. Total Environ. 2021, 784, 146950. [Google Scholar] [CrossRef]
- Miyashita, N.T. Contrasting soil bacterial community structure between the phyla Acidobacteria and Proteobacteria in tropical Southeast Asian and temperate Japanese forests. Genes Genet. Syst. 2015, 90, 61–77. [Google Scholar] [CrossRef] [Green Version]
- Chernov, T.I.; Zhelezova, A.D.; Tkhakakhova, A.K.; Bgazhba, N.A.; Zverev, A.O. Microbiomes of Virgin Soils of Southern Vietnam Tropical Forests. Microbiology 2019, 88, 489–498. [Google Scholar] [CrossRef]
- Lan, G.; Wu, Z.; Yang, C.; Sun, R.; Chen, B.; Zhang, X. Forest conversion alters the structure and functional processes of tropical forest soil microbial communities. Land Degrad. Dev. 2021, 32, 613–627. [Google Scholar] [CrossRef]
- Peterson, D. Categorization of Orthologous Gene Clusters in 92 Ascomycota Genomes Reveals Functions Important for Fungal Phytopathogenicity. Ph.D. Thesis, Northern Illinois University, Dekalb, IL, USA, 2020. [Google Scholar]
- Harley, J. Fungi in ecosystems. J. Ecol. 1971, 59, 653–668. [Google Scholar] [CrossRef]
- Warcup, J. Studies on Basidiomycetes in soil. Trans. Br. Mycol. Soc. 1959, 42, 45–52. [Google Scholar] [CrossRef]
- Curlevski, N.J.A.; Xu, Z.; Anderson, I.C.; Cairney, J.W.G. Converting Australian tropical rainforest to native Araucariaceae plantations alters soil fungal communities. Soil Biol. Biochem. 2010, 42, 14–20. [Google Scholar] [CrossRef]
- Urbina, H.; Scofield, D.G.; Cafaro, M.; Rosling, A. DNA-metabarcoding uncovers the diversity of soil-inhabiting fungi in the tropical island of Puerto Rico. Mycoscience 2016, 57, 217–227. [Google Scholar] [CrossRef]
- Williams, A.; Langridge, H.; Straathof, A.L.; Muhamadali, H.; Hollywood, K.A.; Goodacre, R.; de Vries, F.T. Root functional traits explain root exudation rate and composition across a range of grassland species. J. Ecol. 2021, 110, 21–33. [Google Scholar] [CrossRef]
- Ballauff, J.; Schneider, D.; Edy, N.; Irawan, B.; Daniel, R.; Polle, A. Shifts in root and soil chemistry drive the assembly of belowground fungal communities in tropical land-use systems. Soil Biol. Biochem. 2021, 154, 108140. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [Green Version]
- Hou, Q.; Wang, W.; Yang, Y.; Hu, J.; Bian, C.; Jin, L.; Li, G.; Xiong, X. Rhizosphere microbial diversity and community dynamics during potato cultivation. Eur. J. Soil Biol. 2020, 98, 103176. [Google Scholar] [CrossRef]
- Hartman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angers, D.A.; Caron, J. Plant-induced changes in soil structure: Processes and feedbacks. Biogeochemistry 1998, 42, 55–72. [Google Scholar] [CrossRef]
- Wipf, D.; Krajinski, F.; van Tuinen, D.; Recorbet, G.; Courty, P.-E. Trading on the arbuscular mycorrhiza market: From arbuscules to common mycorrhizal networks. New Phytol. 2019, 223, 1127–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, N.C.; Gehring, C.A. Chapter 4—Mycorrhizas: Symbiotic Mediators of Rhizosphere and Ecosystem Processes. In The Rhizosphere; Cardon, Z.G., Whitbeck, J.L., Eds.; Academic Press: Burlington, VT, USA, 2007; pp. 73–100. [Google Scholar]
- Smith, M.E.; Douhan, G.W.; Fremier, A.K.; Rizzo, D.M. Are True Multihost Fungi the Exception or the Rule? Dominant Ectomycorrhizal Fungi on Pinus sabiniana Differ from Those on Co-occurring Quercus Species. New Phytol. 2009, 182, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Dong, Y.; Xie, X.; Li, X.; Zhang, X.; Shen, X. Effect of annual variation in soil pH on available soil nutrients in pear orchards. Acta Ecol. Sin. 2011, 31, 212–216. [Google Scholar] [CrossRef]
- McCauley, A.; Jones, C.; Jacobsen, J. Soil pH and organic matter. Nutr. Manag. Modul. 2009, 8, 1–12. [Google Scholar]
- Hartmann, A.; Schmid, M.; Van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil Biol. 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Freschet, G.T.; Valverde-Barrantes, O.J.; Tucker, C.M.; Craine, J.M.; McCormack, M.L.; Violle, C.; Fort, F.; Blackwood, C.B.; Urban-Mead, K.R.; Iversen, C.M.; et al. Climate, soil and plant functional types as drivers of global fine-root trait variation. J. Ecol. 2017, 105, 1182–1196. [Google Scholar] [CrossRef] [Green Version]
- Zhalnina, K.; Dias, R.; de Quadros, P.D.; Davis-Richardson, A.; Camargo, F.A.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 2015, 69, 395–406. [Google Scholar] [CrossRef]
- Flores-Rentería, D.; Sánchez-Gallén, I.; Morales-Rojas, D.; Larsen, J.; Álvarez-Sánchez, J. Changes in the Abundance and Composition of a Microbial Community Associated with Land Use Change in a Mexican Tropical Rain Forest. J. Soil Sci. Plant Nutr. 2020, 20, 1144–1155. [Google Scholar] [CrossRef]
- Ji, C.-J.; Yang, Y.-H.; Han, W.-X.; He, Y.-F.; Smith, J.; Smith, P. Climatic and Edaphic Controls on Soil pH in Alpine Grasslands on the Tibetan Plateau, China: A Quantitative Analysis. Pedosphere 2014, 24, 39–44. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Wu, W.; Liu, H. Factors affecting variations of soil pH in different horizons in hilly regions. PLoS ONE 2019, 14, e0218563. [Google Scholar] [CrossRef] [PubMed]
- Du, E.; Terrer, C.; Pellegrini, A.F.A.; Ahlström, A.; van Lissa, C.J.; Zhao, X.; Xia, N.; Wu, X.; Jackson, R.B. Global patterns of terrestrial nitrogen and phosphorus limitation. Nat. Geosci. 2020, 13, 221–226. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, J.; Ding, X.; He, X.; Zhang, F.; Feng, G. Hyphosphere interactions between an arbuscular mycorrhizal fungus and a phosphate solubilizing bacterium promote phytate mineralization in soil. Soil Biol. 2014, 74, 177–183. [Google Scholar] [CrossRef]
- He, D.; Xiang, X.; He, J.-S.; Wang, C.; Cao, G.; Adams, J.; Chu, H. Composition of the soil fungal community is more sensitive to phosphorus than nitrogen addition in the alpine meadow on the Qinghai-Tibetan Plateau. Biol. Fertil. Soils 2016, 52, 1059–1072. [Google Scholar] [CrossRef]
- Huang, J.; Hu, B.; Qi, K.; Chen, W.; Pang, X.; Bao, W.; Tian, G. Effects of phosphorus addition on soil microbial biomass and community composition in a subalpine spruce plantation. Eur. J. Soil Biol. 2016, 72, 35–41. [Google Scholar] [CrossRef]
- Cai, Z.-Q.; Zhang, Y.-H.; Yang, C.; Wang, S. Land-use type strongly shapes community composition, but not always diversity of soil microbes in tropical China. CATENA 2018, 165, 369–380. [Google Scholar] [CrossRef]
- Magan, N. Fungi in extreme environments. Mycota 2007, 4, 85–103. [Google Scholar]
- Chen, H.; Mothapo, N.V.; Shi, W. Soil moisture and pH control relative contributions of fungi and bacteria to N2O production. Microb. Ecol. 2015, 69, 180–191. [Google Scholar] [CrossRef]
- Venkateswarlu, B.; Rao, A.; Raina, P. Evaluation of phosphorus solubilisation by microorganisms isolated from Aridisols. J. Indian Soc. Soil Sci. 1984, 32, 273–277. [Google Scholar]
- Li, J.; Li, Z.; Wang, F.; Zou, B.; Chen, Y.; Zhao, J.; Mo, Q.; Li, Y.; Li, X.; Xia, H. Effects of nitrogen and phosphorus addition on soil microbial community in a secondary tropical forest of China. Biol. Fertil. Soils 2015, 51, 207–215. [Google Scholar] [CrossRef]
- Wang, J.; Liu, G.; Zhang, C.; Wang, G.; Fang, L.; Cui, Y. Higher temporal turnover of soil fungi than bacteria during long-term secondary succession in a semiarid abandoned farmland. Soil Tillage Res. 2019, 194, 104305. [Google Scholar] [CrossRef]


| Plant Species | Plant Life Forms | Plant Categories | DBH 1 (cm) | Height 1 (m) |
|---|---|---|---|---|
| Michelia mediocris Dandy | Tree | Angiosperm | 6.22 ± 5.31 | 5.76 ± 2.43 |
| Podocarpus neriifolius D. Don | Tree | Gymnosperm | 5.62 ± 5.19 | 4.600 ± 1.39 |
| Syzygium buxifolium Hook. et Arn. | Shrub or small tree | Angiosperm | 8.02 ± 3.29 | 5.560 ± 1.13 |
| Cyclobalanopsis disciformis (Chun et Tsiang) Y. C. Hsu et H. W. Jen | Tree | Angiosperm | 7.62 ± 11.45 | 4.18 ± 2.20 |
| Manglietia fordiana var. hainanensis (Dandy) N. H. Xia | Tree | Angiosperm | 1.76 ± 0.53 | 2.90 ± 1.25 |
| Pinus fenzeliana Hand.-Mzt. | Tree | Gymnosperm | 36.24 ± 16.41 | 12.14 ± 5.21 |
| Castanopsis faberi Hance | Tree | Angiosperm | 2.63 ± 1.79 | 2.50 ± 0.87 |
| Osmanthus didymopetalus P. S. Green | Tree | Angiosperm | 2.92 ± 2.56 | 4.12 ± 1.89 |
| Distylium racemosum Sieb. et Zucc. | Shrub or small tree | Angiosperm | 16.46 ± 4.17 | 9.00 ± 1.00 |
| Allomorphia balansae Cogn. | Shrub | Angiosperm | 2.44 ± 1.45 | 2.75 ± 0.60 |
| Olea dioica Roxb. | Shrub or small tree | Angiosperm | 2.17 ± 1.10 | 2.93 ± 0.66 |
| Syzygium championii (Benth.) Merr. et Perry | Shrub to small tree | Angiosperm | 2.54 ± 1.59 | 3.820 ± 2.27 |
| Melastoma penicillatum Naud. | Shrub | Angiosperm | 2.13 ± 0.79 | 3.00 ± 0.84 |
| Traits | Abbreviation | Unit | Ecological Strategies | |
|---|---|---|---|---|
| Morphology traits | Specific root length | SRL | cm/g | Resource acquisition. |
| Root tissue density | RTD | g/cm3 | Transport, support and defense. | |
| Specific root area | SRA | cm2/g | Resource acquisition and defense. | |
| Chemical traits | Root carbon content | RC | g/kg | Microbial carbon source. |
| Root nitrogen content | RN | g/kg | Microbial nutrient source. | |
| Root phosphorus content | RP | g/kg | Microbial nutrient source. |
| RC | RN | RP | RTD | SRL | SRA | pH | SOM | STN | STP | SAN | SAP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | OTUs | 0.55 | 0.59 | 0.04 * | 0.23 | 0.26 | 0.15 | 0.009 ** | 0.85 | 0.94 | 0.67 | 0.16 | 0.28 |
| Chao1 | 0.56 | 0.57 | 0.04 * | 0.22 | 0.28 | 0.16 | 0.01 * | 0.86 | 0.96 | 0.66 | 0.18 | 0.31 | |
| Faith-pd | 0.89 | 0.33 | 0.04 * | 0.35 | 0.04 * | 0.04 * | 0.0002 *** | 0.79 | 0.44 | 0.43 | 0.06 | 0.12 | |
| Shannon | 0.34 | 0.69 | 0.16 | 0.32 | 0.25 | 0.16 | 0.002 ** | 0.85 | 0.82 | 0.72 | 0.31 | 0.08 | |
| Simpson | 0.40 | 0.60 | 0.31 | 0.60 | 0.22 | 0.19 | 0.004 ** | 0.89 | 0.93 | 0.84 | 0.83 | 0.03 * | |
| Fungi | OTUs | 1.00 | 0.23 | 0.47 | 0.97 | 0.57 | 0.56 | 0.001 ** | 0.62 | 0.44 | 0.37 | 0.58 | 0.05 |
| Chao1 | 1.00 | 0.23 | 0.47 | 0.97 | 0.57 | 0.56 | 0.001 ** | 0.62 | 0.44 | 0.37 | 0.58 | 0.05 | |
| Faith-pd | 0.90 | 0.39 | 0.47 | 0.88 | 0.43 | 0.47 | 0.0002 *** | 0.40 | 0.19 | 0.17 | 0.99 | 0.03 * | |
| Shannon | 0.69 | 0.18 | 0.85 | 0.31 | 0.56 | 0.33 | 0.02 * | 0.87 | 0.53 | 0.79 | 0.10 | 0.19 | |
| Simpson | 0.89 | 0.21 | 0.60 | 0.25 | 0.44 | 0.21 | 0.06 | 0.75 | 0.46 | 0.86 | 0.18 | 0.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Z.; Wang, Y.; Xiao, C.; Zhang, D.; Feng, G.; Long, W. Effects of Plant Fine Root Functional Traits and Soil Nutrients on the Diversity of Rhizosphere Microbial Communities in Tropical Cloud Forests in a Dry Season. Forests 2022, 13, 421. https://doi.org/10.3390/f13030421
Deng Z, Wang Y, Xiao C, Zhang D, Feng G, Long W. Effects of Plant Fine Root Functional Traits and Soil Nutrients on the Diversity of Rhizosphere Microbial Communities in Tropical Cloud Forests in a Dry Season. Forests. 2022; 13(3):421. https://doi.org/10.3390/f13030421
Chicago/Turabian StyleDeng, Zhiyan, Yichen Wang, Chuchu Xiao, Dexu Zhang, Guang Feng, and Wenxing Long. 2022. "Effects of Plant Fine Root Functional Traits and Soil Nutrients on the Diversity of Rhizosphere Microbial Communities in Tropical Cloud Forests in a Dry Season" Forests 13, no. 3: 421. https://doi.org/10.3390/f13030421





