Plant Biodiversity Homogenization across the Chronosequence in Highly Fragmented Landscapes in the Colombian Andean–Amazonian Transition
Abstract
:1. Introduction
2. Materials and Methods
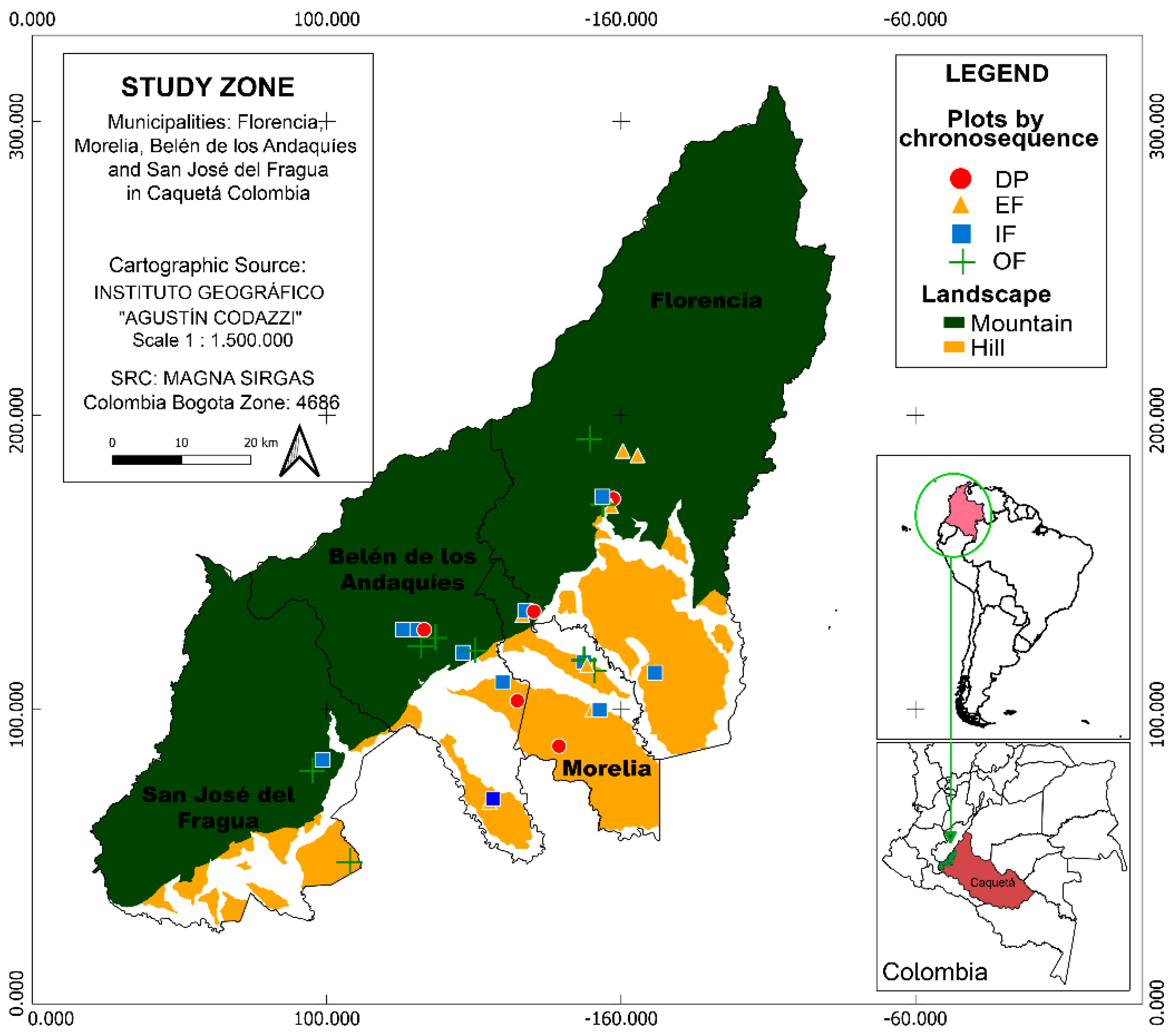
2.1. Study Area and Sampling Design
2.2. Floristic Data Collection
2.3. Environmental Parameters
2.4. Data Analysis
3. Results
3.1. Plant Community Composition and Growth Habits
3.2. Species Richness Accumulation
3.3. Indicator Species Identification
3.4. Plant Community Composition Dissimilarity
3.5. Influence of Environmental Filters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. FAO Global Forest Resources Assessment (FRA) 2020; FAO: Rome, Italy, 2020. [Google Scholar]
- Pain, A.; Marquardt, K.; Lindh, A.; Hasselquist, N.J. What Is Secondary about Secondary Tropical Forest? Rethinking Forest Landscapes. Hum. Ecol. 2020, 49, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Chazdon, R.L.; Peres, C.; Dent, D.; Sheil, D.; Lugo, A.E.; Lamb, D.; Stork, N.; Miller, S. The Potential for Species Conservation in Tropical Secondary Forests. Conserv. Biol. 2009, 23, 1406–1417. [Google Scholar] [CrossRef] [PubMed]
- Newbold, T.; Hudson, L.N.; Hill, S.L.L.; Contu, S.; Lysenko, I.; Senior, R.A.; Börger, L.; Bennett, D.J.; Choimes, A.; Collen, B.; et al. Global effects of land use on local terrestrial biodiversity. Nature 2015, 520, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Rozendaal, D.M.A.; Bongers, F.; Aide, T.M.; Alvarez-Dávila, E.; Ascarrunz, N.; Balvanera, P.; Becknell, J.M.; Bentos, T.V.; Brancalion, P.H.S.; Cabral, G.A.L.; et al. Biodiversity recovery of Neotropical secondary forests. Sci. Adv. 2019, 5, eaau3114. [Google Scholar] [CrossRef]
- Martinez-Garza, C.; Howe, H.F. Restoring tropical diversity: Beating the time tax on species loss. J. Appl. Ecol. 2003, 40, 423–429. [Google Scholar] [CrossRef]
- Poorter, L.; Craven, D.; Jakovac, C.C.; van der Sande, M.T.; Amissah, L.; Bongers, F.; Chazdon, R.L.; Farrior, C.E.; Kambach, S.; Meave, J.A.; et al. Multidimensional tropical forest recovery. Science 2021, 374, 1370–1376. [Google Scholar] [CrossRef]
- DeWalt, S.J.; Maliakal, S.K.; Denslow, J.S. Changes in vegetation structure and composition along a tropical forest chronosequence: Implications for wildlife. For. Ecol. Manag. 2003, 182, 139–151. [Google Scholar] [CrossRef]
- Phillips, H.R.P.; Newbold, T.; Purvis, A. Land-use effects on local biodiversity in tropical forests vary between continents. Biodivers. Conserv. 2017, 26, 2251–2270. [Google Scholar] [CrossRef]
- Olden, J.D.; Rooney, T.P. On defining and quantifying biotic homogenization. Glob. Ecol. Biogeogr. 2006, 15, 113–120. [Google Scholar] [CrossRef]
- Baeten, L.; Vangansbeke, P.; Hermy, M.; Peterken, G.; Vanhuyse, K.; Verheyen, K. Distinguishing between turnover and nestedness in the quantification of biotic homogenization. Biodivers. Conserv. 2012, 21, 1399–1409. [Google Scholar] [CrossRef] [Green Version]
- Olden, J.D.; Poff, N.L. Redundancy and the choice of hydrologic indices for characterizing streamflow regimes. River Res. Appl. 2003, 19, 101–121. [Google Scholar] [CrossRef]
- Lewis, S.L.; Maslin, M.A. Defining the Anthropocene. Nature 2015, 519, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Van Der Plas, F.; Manning, P.; Soliveres, S.; Allan, E.; Scherer-Lorenzen, M.; Verheyen, K.; Wirth, C.; Zavala, M.A.; Ampoorter, E.; Baeten, L.; et al. Biotic homogenization can decrease landscape-scale forest multifunctionality. Proc. Natl. Acad. Sci. USA 2016, 113, 3557–3562. [Google Scholar] [CrossRef] [PubMed]
- Baiser, B.; Olden, J.; Record, S.; Lockwood, J.; McKinney, M.L. Pattern and process of biotic homogenization in the New Pangaea. Proc. R. Soc. B Biol. Sci. 2012, 279, 4772–4777. [Google Scholar] [CrossRef]
- Smith, C.C.; Healey, J.R.; Berenguer, E.; Young, P.J.; Taylor, B.; Elias, F.; Espírito-Santo, F.; Barlow, J. Old-growth forest loss and secondary forest recovery across Amazonian countries. Environ. Res. Lett. 2021, 16, 085009. [Google Scholar] [CrossRef]
- Martínez-Ramos, M.; Barragán, F.; Mora, F.; Maza-Villalobos, S.; Arreola-Villa, L.F.; Bhaskar, R.; Bongers, F.; Lemus-Herrera, C.; Paz, H.; Martínez-Yrizar, A.; et al. Differential ecological filtering across life cycle stages drive old-field succession in a neotropical dry forest. For. Ecol. Manag. 2020, 482, 118810. [Google Scholar] [CrossRef]
- Chazdon, R.L. Tropical Forest Regeneration☆. In Reference Module in Life Sciences; Elsevier: London, UK, 2017. [Google Scholar]
- IDEAM—Instituto de Hidrología, Meteorología y Estudios Ambientales. Presentación balance deforestación 2019 (1); IDEAM: Bogotá, Colombia, 2019.
- Gloor, M.; Barichivich, J.; Ziv, G.; Brienen, R.J.W.; Schongart, J.; Peylin, P.; Cintra, B.B.L.; Feldpausch, T.R.; Phillips, O.L.; Baker, J.W. Recent Amazon climate as background for possible ongoing and future changes of Amazon humid forests. Glob. Biogeochem. Cycles 2015, 29, 1384–1399. [Google Scholar] [CrossRef]
- IDEAM—Instituto de Hidrología, Meteorología y Estudios Ambientales. Resultados Del Monitoreo de Deforestación: 1. Año 2020. 2. Primer Trimestre Año 2021; IDEAM: Bogotá, Colombia, 2020.
- Rodríguez-León, C.H.; Peña-Venegas, C.P.; Sterling, A.; Muñoz-Ramirez, H.; Virguez-Díaz, Y.R. Changes in Soil-Borne Communities of Arbuscular Mycorrhizal Fungi during Natural Regrowth of Abandoned Cattle Pastures Are Indicative of Ecosystem Restoration. Agronomy 2021, 11, 2468. [Google Scholar] [CrossRef]
- Rodríguez-León, C.H.; Peña-Venegas, C.P.; Sterling, A.; Castro, D.; Mahecha-Virguez, L.K.; Virguez-Díaz, Y.R.; Silva-Olaya, A.M. Soil Quality Restoration during the Natural Succession of Abandoned Cattle Pastures in Deforested Landscapes in the Colombian Amazon. Agronomy 2021, 11, 2484. [Google Scholar] [CrossRef]
- Pires, A.P.F.; Srivastava, D.S.; Farjalla, V.F. Is Biodiversity Able to Buffer Ecosystems from Climate Change? What We Know and What We Don’t. BioScience 2018, 68, 273–280. [Google Scholar] [CrossRef]
- Chazdon, R. Second Growth. The Promise of Tropical Forest Regeneration in an Age of Deforestation. Available online: https://www.degruyter.com/document/doi/10.7208/9780226118109/html?lang=en (accessed on 12 June 2022).
- Ferguson, B.G.; Vandermeer, J.; Morales, H.; Griffith, D.M. Post-Agricultural Succession in El Petén, Guatemala. Conserv. Biol. 2003, 17, 818–828. [Google Scholar] [CrossRef]
- Uriarte, M.; Condit, R.; Canham, C.; Hubbell, S.P. A spatially explicit model of sapling growth in a tropical forest: Does the identity of neighbours matter? J. Ecol. 2004, 92, 348–360. [Google Scholar] [CrossRef]
- Comita, L.S.; Thompson, J.; Uriarte, M.; Jonckheere, I.; Canham, C.; Zimmerman, J.K. Interactive effects of land use history and natural disturbance on seedling dynamics in a subtropical forest. Ecol. Appl. 2010, 20, 1270–1284. [Google Scholar] [CrossRef] [PubMed]
- Instituto Geográfico Agustin Codazzi (IGAC). Caquetá, Características Geográficas; Imprenta Nacional de Colombia: Bogotá, Colombia, 2010. [Google Scholar]
- Abbas, S.; Nichol, J.E.; Zhang, J.; Fischer, G.A. The accumulation of species and recovery of species composition along a 70 year succession in a tropical secondary forest. Ecol. Indic. 2019, 106, 105524. [Google Scholar] [CrossRef]
- Hu, Y.-K.; Pan, X.; Liu, X.-Y.; Fu, Z.-X.; Zhang, M.-Y. Above-and Belowground Plant Functional Composition Show Similar Changes during Temperate Forest Swamp Succession. Front. Plant Sci. 2021, 12, 658883. [Google Scholar] [CrossRef]
- Rocha-Ortega, M.; García-Martínez, M. Importance of Nesting Resources and Soil Conditions for the Recovery of Ant Diversity During Secondary Succession in a Tropical Rainforest. Trop. Conserv. Sci. 2018, 11, 1940082918787063. [Google Scholar] [CrossRef]
- Zambiazi, D.C.; Fantini, A.C.; Piotto, D.; Siminski, A.; Vibrans, A.C.; Oller, D.C.; Piazza, G.E.; Peña-Claros, M. Timber stock recovery in a chronosequence of secondary forests in Southern Brazil: Adding value to restored landscapes. For. Ecol. Manag. 2021, 495, 119352. [Google Scholar] [CrossRef]
- Teixeira, H.M.; Cardoso, I.M.; Bianchi, F.J.; Silva, A.D.C.; Jamme, D.; Peña-Claros, M. Linking vegetation and soil functions during secondary forest succession in the Atlantic forest. For. Ecol. Manag. 2019, 457, 117696. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef]
- Velasquez, E.; Lavelle, P.; Andrade, M. GISQ, a multifunctional indicator of soil quality. Soil Biol. Biochem. 2007, 39, 3066–3080. [Google Scholar] [CrossRef]
- McGlinn, D.; Xiao, X.; McGill, B.; May, F.; Engel, T.; Oliver, C.; Blowes, S.; Knight, T.; Purschke, O.; Gotelli, N.; et al. Package ‘Mobr’: Measurement of Biodiversity Package Version 2.0.2; The Comprehensive R Archive Network: Vienna, Austria, 2021. [Google Scholar]
- Chao, A. Estimating the Population Size for Capture-Recapture Data with Unequal Catchability. Biometrics 1987, 43, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Chiarucci, A.; Bacaro, G.; Rocchini, D.; Ricotta, C.; Palmer, M.; Scheiner, S. Spatially constrained rarefaction: Incorporating the autocorrelated structure of biological communities into sample-based rarefaction. Community Ecol. 2009, 10, 209–214. [Google Scholar] [CrossRef]
- Roberts, D.W. Package: “Labdsv”: Ordination and Multivariate Analysis for Ecology Package Version 2.0-1; The Comprehensive R Archive Network: Vienna, Austria, 2019. [Google Scholar]
- Oksanen, J.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; et al. Package ‘Vegan’: Community Ecology Package Version 2.5-7; The Comprehensive R Archive Network: Vienna, Austria, 2018. [Google Scholar]
- Anderson, M.J. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Martinez-Arbizu, P. Package: “PairwiseAdonis”: Pairwise Multilevel Comparison Using Adonis Package Version: 0.0.1; The Comprehensive R Archive Network: Vienna, Austria, 2017. [Google Scholar]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier B.V.: Oxford, UK, 2012. [Google Scholar]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R, 2nd ed.; Springer International Publishing AG: Cham, Switzerland, 2018. [Google Scholar]
- McArdle, B.H.; Anderson, M.J. Fitting Multivariate Models to Community Data: A Comment on Distance-Based Redundancy Analysis. Ecology 2001, 82, 290–297. [Google Scholar] [CrossRef]
- Borcard, D.; Legendre, P. All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecol. Model. 2002, 153, 51–68. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.-B.; Thioulouse, J. Ade4: Analysis of Ecological Data: Exploratory and Euclidean Methods in Environmental Sciences, R Package Version 1.7-16; The Comprehensive R Archive Network: Vienna, Austria, 2020. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing v. 4.0.3; The Comprehensive R Archive Network: Vienna, Austria, 2020. [Google Scholar]
- RStudio Team. RStudio v.1.3.1093; RStudi—Open Source & Professional Software for Data Science: Boston, MA, USA, 2020. [Google Scholar]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, C.W. InfoStat v. 2020; Universidad Nacional de Córdoba: Córdoba, Argentina, 2020. [Google Scholar]
- Duivenvoorden, J.F. Vascular plant species counts in the rain forests of the middle Caquetá area, Colombian Amazonia. Biodivers. Conserv. 1994, 3, 685–715. [Google Scholar] [CrossRef]
- Duque, A.; Sánchez, M.; Cavelier, J.; Duivenvoorden, J.F. Different floristic patterns of woody understorey and canopy plants in Colombian Amazonia. J. Trop. Ecol. 2002, 18, 499–525. [Google Scholar] [CrossRef]
- Olden, J.D. Biotic Homogenization. In eLS.; Wiley: Chichester, UK, 2008. [Google Scholar]
- Calle, H.; Flórez, J. Así Funciona El Tráfico de Madera en Colombia. Available online: https://es.mongabay.com/2018/10/trafico-de-madera-en-colombia-amazonia-bosques/ (accessed on 14 June 2022).
- Keith, S.A.; Newton, A.C.; Morecroft, M.D.; Bealey, C.E.; Bullock, J.M. Taxonomic homogenization of woodland plant communities over 70 years. Proc. R. Soc. B Biol. Sci. 2009, 276, 3539–3544. [Google Scholar] [CrossRef] [Green Version]
- Daru, B.H.; Davies, T.J.; Willis, C.G.; Meineke, E.K.; Ronk, A.; Zobel, M.; Pärtel, M.; Antonelli, A.; Davis, C.C. Widespread homogenization of plant communities in the Anthropocene. Nat. Commun. 2021, 12, 6983. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Koyanagi, T.F.; Matsumura, T.; Koyama, A. Patterns of plant diversity loss and species turnover resulting from land abandonment and intensification in semi-natural grasslands. J. Environ. Manag. 2018, 218, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Clements, F.E. Plant Succession, An Analysis of the Devel-Opment of Vegetation; Carnegie Institution of Washington: Washington, DC, USA, 1916. [Google Scholar]
- Arroyo-Rodríguez, V.; Rös, M.; Escobar, F.; Melo, F.P.L.; Santos, B.A.; Tabarelli, M.; Chazdon, R. Plant β-diversity in fragmented rain forests: Testing floristic homogenization and differentiation hypotheses. J. Ecol. 2013, 101, 1449–1458. [Google Scholar] [CrossRef]
- Rodríguez, C.; Sterling, A. Sucesión Ecológica y Restauración En Paisajes Fragmentados de La Amazonia Colombiana. Tomo 1. Composición, Estructura y Función En La Sucesión Secundaria; Instituto Amazónico de Investigaciones Científicas-SINCHI: Bogotá, Colombia, 2020. [Google Scholar]
- Martínez, L.; Zinck, J. Temporal variation of soil compaction and deterioration of soil quality in pasture areas of Colombian Amazonia. Soil Tillage Res. 2004, 75, 3–18. [Google Scholar] [CrossRef]








| Chronosequence | Age Range (Years) | Age Midpoint (Years) | Landscape | Number of Plots |
|---|---|---|---|---|
| Degraded pasture (DP) | <3 | 1.5 | Hill | 2 |
| Mountain | 3 | |||
| Early forest (EF) | 10–20 | 15 | Hill | 3 |
| Mountain | 4 | |||
| Intermediate forest (IF) | 25–40 | 32.5 | Hill | 6 |
| Mountain | 6 | |||
| Old-growth forest or mature forest (OF) | >90 | 90 | Hill | 3 |
| Mountain | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-León, C.H.; Roa-Fuentes, L.L.; Sterling, A.; Suárez, J.C. Plant Biodiversity Homogenization across the Chronosequence in Highly Fragmented Landscapes in the Colombian Andean–Amazonian Transition. Forests 2022, 13, 1422. https://doi.org/10.3390/f13091422
Rodríguez-León CH, Roa-Fuentes LL, Sterling A, Suárez JC. Plant Biodiversity Homogenization across the Chronosequence in Highly Fragmented Landscapes in the Colombian Andean–Amazonian Transition. Forests. 2022; 13(9):1422. https://doi.org/10.3390/f13091422
Chicago/Turabian StyleRodríguez-León, Carlos H., Lilia L. Roa-Fuentes, Armando Sterling, and Juan Carlos Suárez. 2022. "Plant Biodiversity Homogenization across the Chronosequence in Highly Fragmented Landscapes in the Colombian Andean–Amazonian Transition" Forests 13, no. 9: 1422. https://doi.org/10.3390/f13091422
APA StyleRodríguez-León, C. H., Roa-Fuentes, L. L., Sterling, A., & Suárez, J. C. (2022). Plant Biodiversity Homogenization across the Chronosequence in Highly Fragmented Landscapes in the Colombian Andean–Amazonian Transition. Forests, 13(9), 1422. https://doi.org/10.3390/f13091422





