Construction of SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Growth Traits in Eucommia ulmoides Oliver
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. DNA Library Preparation and High-Throughput Sequencing
2.3. SNP Calling and Genotyping
2.4. Growth Traits and Statistical Analysis
2.5. High-Density Genetic Map Construction and QTL Analysis
2.6. Prediction of Candidate Genes and Functional Annotation
3. Results
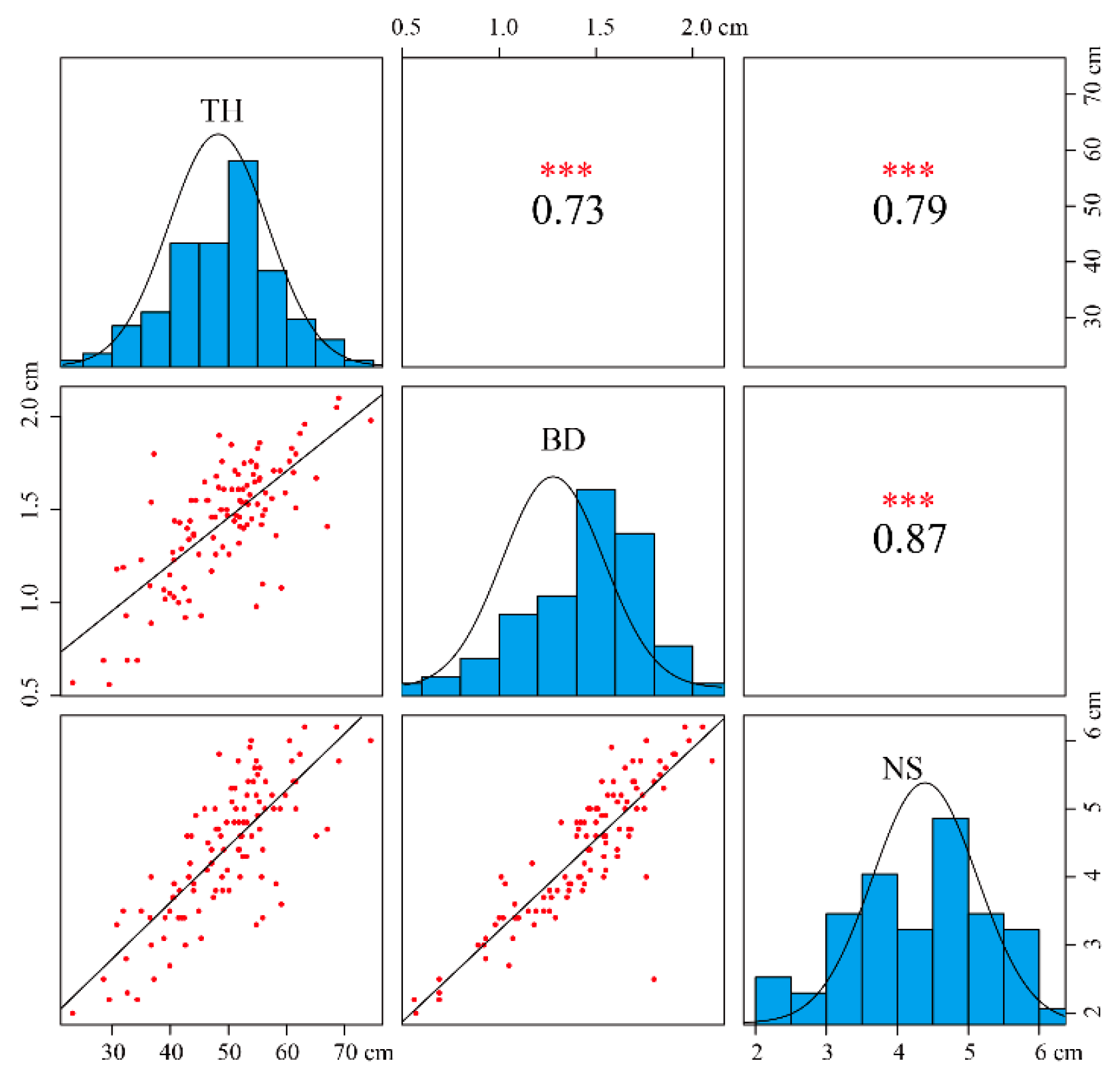
3.1. Analysis of Growth Traits
3.2. GBS Sequencing Results and Analysis
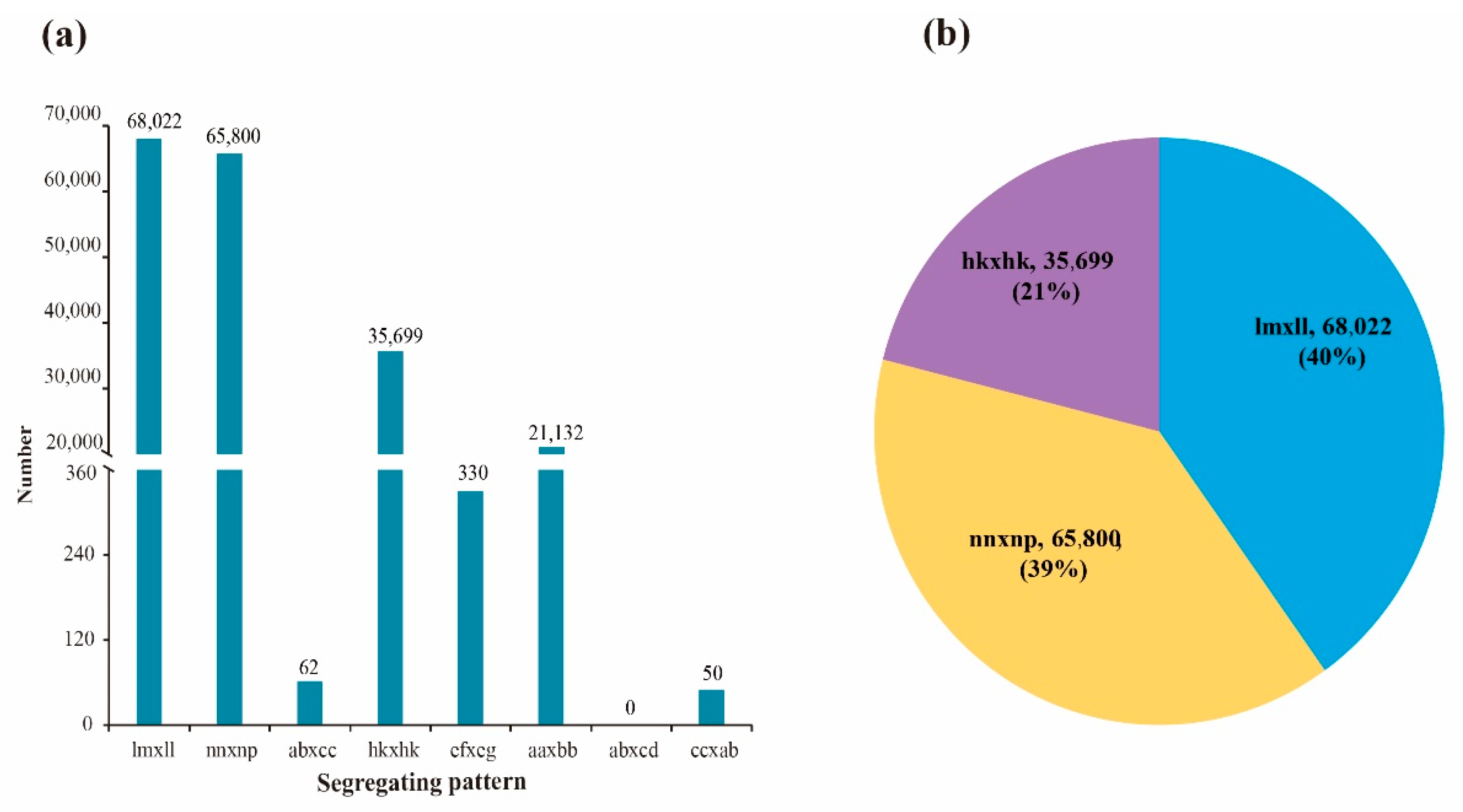
3.3. SNP Marker Detection and Genotyping
3.4. High-Density Genetic Map Construction
3.5. QTL Mapping of Growth Traits
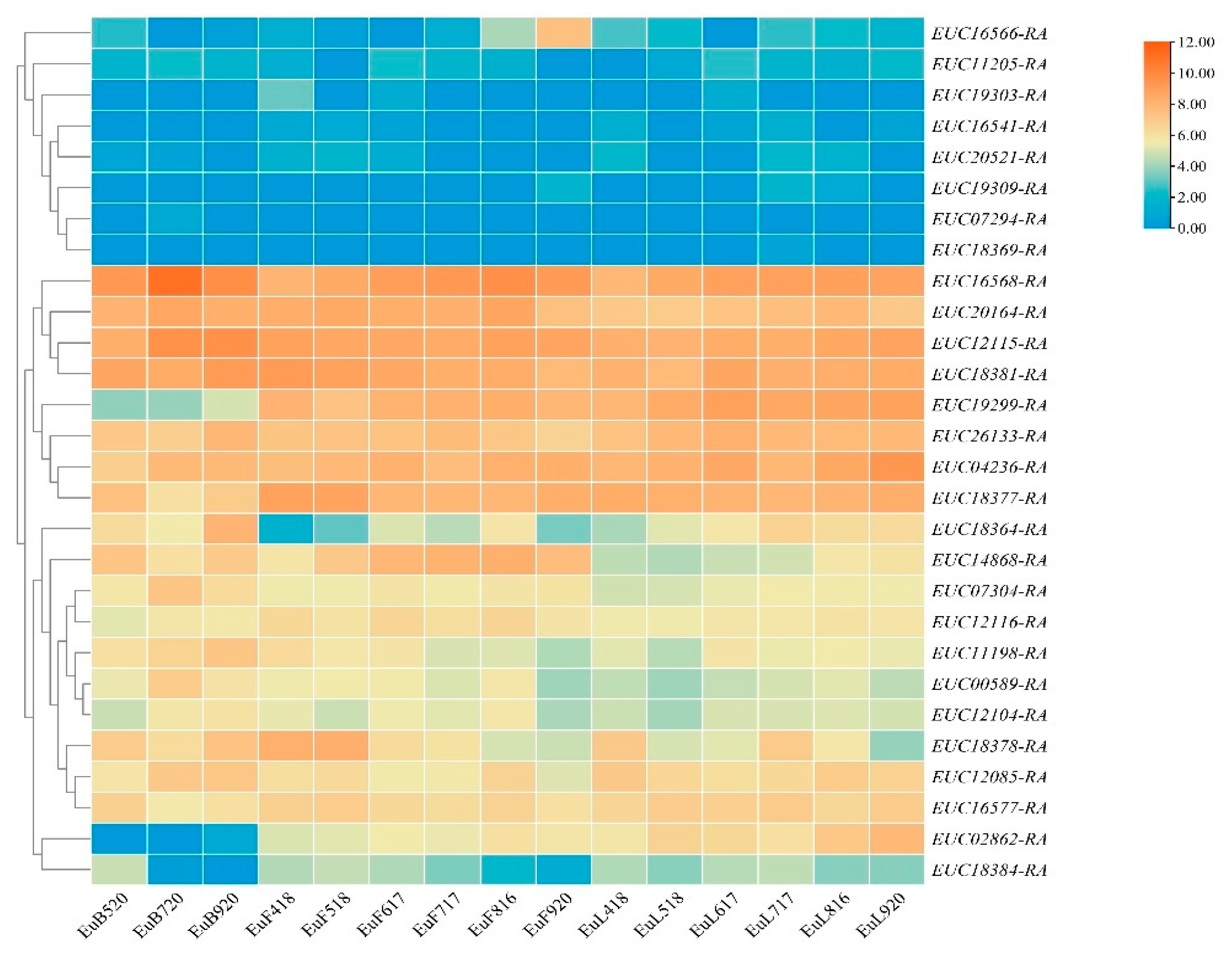
3.6. Expression and Annotation of the Candidate Genes
4. Discussion
4.1. Population Selection for Map Construction
4.2. The Advantage of SNP-Marker and GBS Technology
4.3. High-Density Genetic Maps
4.4. QTL Mapping and Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wuyun, T.N.; Wang, L.; Liu, H.; Wang, X.; Zhang, L.; Bennetzen, J.L.; Li, T.; Yang, L.; Liu, P.; Du, L. The Hardy Rubber Tree Genome Provides Insights into the Evolution of Polyisoprene Biosynthesis. Mol. Plant. 2018, 11, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Call, V.B.; Dilcher, D.L. The fossil record of Eucommia (Eucommiaceae) in North America. Am. J. Bot. 1997, 84, 798–814. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Li, Z.; Li, Y.; Wang, S.; Li, L.; Liu, M. Update of Genetic Linkage Map and QTL Analysis for Growth Traits in Eucommia ulmoides Oliver. Forests 2020, 11, 311. [Google Scholar] [CrossRef]
- Du, H. China Eucommia Pictorial; China Forestry Publishing House: Beijing, China, 2014. [Google Scholar]
- Du, H.; Hu, W.; Yu, R. The Report on Development of China’s Eucommia Rubber Resources and Industry (2014–2015); Social Sciences Academic Press: Beijing, China, 2015. [Google Scholar]
- Nakazawa, Y.; Bamba, T.; Takeda, T.; Uefuji, H.; Harada, Y.; Li, X.; Chen, R.; Inoue, S.; Tutumi, M.; Shimizu, T. Production of Eucommia-rubber from Eucommia ulmoides Oliv. (Hardy Rubber Tree). Plant Biotechnol. 2009, 26, 71–79. [Google Scholar] [CrossRef]
- Wang, C.; Tang, L.; He, J.; Li, J.; Wang, Y. Ethnobotany, Phytochemistry and Pharmacological Properties of Eucommia ulmoides: A Review. Am. J. Chin. Med. 2019, 47, 259–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, L.; Li, L.; Zhou, Q.; Li, Y.; Li, J.; Wang, Y. Geographic Authentication of Eucommia ulmoides Leaves Using Multivariate Analysis and Preliminary Study on the Compositional Response to Environment. Front. Plant Sci. 2020, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, D.; Li, Z.; Wei, J.; Jin, C.; Liu, M. A Molecular Genetic Linkage Map of Eucommia ulmoides and Quantitative Trait Loci (QTL) Analysis for Growth Traits. Int. J. Mol. Sci. 2014, 15, 2053–2074. [Google Scholar] [CrossRef]
- He, D.; Lin, Z.; Zhang, X.; Nie, Y.; Guo, X.; Zhang, Y.; Li, W. QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × Gossypium barbadense. Euphytica 2006, 153, 181–197. [Google Scholar] [CrossRef]
- Fang, X.; Dong, K.; Wang, X.; Liu, T.; He, J.; Ren, R.; Zhang, L.; Liu, R.; Liu, X.; Li, M. A high density genetic map and QTL for agronomic and yield traits in Foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genom. 2016, 17, 336. [Google Scholar] [CrossRef]
- Adak, A.; Conrad, C.; Chen, Y.; Wilde, S.C.; Murray, S.C.; AndersonII, S.L.; Subramanian, N.K. Validation of functional polymorphisms affecting maize plant height by unoccupied aerial systems discovers novel temporal phenotypes. G3 Genes Genomes Genet. 2021, 11, jkab075. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Li, L.; Wei, Y.; Li, Z. The first genetic linkage map of Eucommia ulmoides. J. Genet. 2014, 93, 13–20. [Google Scholar] [CrossRef]
- Shavrukov, Y. Comparison of SNP and CAPS markers application in genetic research in wheat and barley. BMC Plant Biol. 2016, 16, 11. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef]
- Zhang, Z.; Wei, T.; Zhong, Y.; Li, X.; Huang, J. Construction of a high-density genetic map of Ziziphus jujuba Mill. using genotyping by sequencing technology. Tree Genet. Genomes 2016, 12, 76. [Google Scholar] [CrossRef]
- Wang, L.; Du, H.; Li, T.; Wuyun, T.N. De novo transcriptome sequencing and identification of genes related to salt stress in Eucommia ulmoides Oliver. Trees 2018, 32, 151–163. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.T.; Fennell, J.; Ruan, N.; Homer, G.; Marth, G.; Abecasis, R. Durbin: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Ooijen, J. JoinMap 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Population; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Xu, S. Quantitative trait locus mapping can benefit from segregation distortion. Genetics 2008, 180, 2201–2208. [Google Scholar] [CrossRef]
- Ooijen, J.V.; Ooijen, J.; Hoorn, J.; Duin, J.; Van, J.W. MapQTL®6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma BV: Wageningen, The Netherlands, 2009. [Google Scholar]
- Wang, Z.; Zhang, Z.; Tang, H.; Zhang, Q.; Zhou, G.; Li, X. High-Density Genetic Map Construction and QTL Mapping of Leaf and Needling Traits in Ziziphus jujuba Mill. Front. Plant Sci. 2019, 10, 1424. [Google Scholar] [CrossRef]
- Schmid, R.; Blaxter, M.L. annot8r: GO, EC and KEGG annotation of EST datasets. BMC Bioinform. 2008, 9, 180. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Salazar, J.A.; Pacheco, I.; Shinya, P.; Zapata, P.; Silva, C.; Aradhya, M.; Velasco, D.; Ruiz, D.; Martínez-Gómez, P.; Infante, R. Genotyping by Sequencing for SNP-Based Linkage Analysis and Identification of QTLs Linked to Fruit Quality Traits in Japanese Plum (Prunus salicina Lindl.). Front. Plant Sci. 2017, 8, 476. [Google Scholar] [CrossRef]
- Hu, M.; Zhang, H.; Wang, B.; Song, Z.; Gao, Y.; Yuan, C.; Huang, C.; Zhao, L.; Zhang, Y.; Wang, L. Transcriptomic analysis provides insights into the AUXIN RESPONSE FACTOR 6-mediated repression of nicotine biosynthesis in tobacco (Nicotiana tabacum L.). Plant Mol. Biol. 2021, 107, 21–36. [Google Scholar] [CrossRef]
- Huang, G.; Hu, H.; Meene, A.; Zhang, J.; Dong, L.; Zheng, S.; Zhang, F.; Betts, N.S.; Liang, W.; Bennett, M.J. AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 2021, 33, 3120–3133. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant-Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Y.; Guo, Y.; Su, K.; Shi, X.; Liu, D.; Zhang, J. High-density genetic linkage-map construction of hawthorn and QTL mapping for important fruit traits. PLoS ONE 2020, 15, e0229020. [Google Scholar] [CrossRef] [Green Version]
- Wang, M. Forest Genetic and Breeding; China Forestry Publishing House: Beijing, China, 2001. [Google Scholar]
- Lu, N.; Zhang, M.; Xiao, Y.; Han, D.; Liu, Y.; Zhang, Y.; Yi, F.; Zhu, T.; Ma, W.; Fan, E. Construction of a high-density genetic map and QTL mapping of leaf traits and plant growth in an interspecific F1 population of Catalpa bungei × Catalpa duclouxii Dode. BMC Plant Biol. 2019, 19, 596. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, S.; Gan, Z.; Zeng, R.; Zhang, J.; Hu, C. High-Density Genetic Map Construction and Identification of QTLs Controlling Leaf Abscission Trait in Poncirus trifoliata. Int. J. Mol. Sci. 2021, 22, 5723. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Koch, J.; Coggeshall, M.; Carlson, J. The first genetic linkage map for Fraxinus pennsylvanica and syntenic relationships with four related species. Plant Mol. Biol. 2019, 99, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Guo, Y.; Su, K.; Liu, Z.; Ren, Z.; Li, K.; Guo, X. Construction of a highly saturated Genetic Map for Vitis by Next-generation Restriction Site-associated DNA Sequencing. BMC Plant Biol. 2018, 18, 347. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yuan, W.; Dong, M.; Han, Y.; Shang, F. The First Genetic Map in Sweet Osmanthus (Osmanthus fragrans Lour.) Using Specific Locus Amplified Fragment Sequencing. Front. Plant Sci. 2017, 8, 1621. [Google Scholar] [CrossRef] [PubMed]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, J.A. Novel genetic mapping tools in plants: SNPs and LD-based approaches. Plant Sci. 2002, 162, 329–333. [Google Scholar] [CrossRef]
- Liu, J.; Huang, S.; Sun, M.; Liu, S.; Liu, Y.; Wang, W.; Zhang, X.; Wang, H.; Hua, W. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 2012, 8, 34. [Google Scholar] [CrossRef]
- Donato, M.D.; Peters, S.O.; Mitchell, S.E.; Hussain, T.; Imumorin, I.G. Genotyping-by-Sequencing (GBS): A Novel, Efficient and Cost-Effective Genotyping Method for Cattle Using Next-Generation Sequencing. PLoS ONE 2013, 8, e62137. [Google Scholar]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef]
- Zhang, K.; Kuraparthy, V.; Fang, H.; Zhu, L.; Sood, S.; Jones, D.C. High-density linkage map construction and QTL analyses for fiber quality, yield and morphological traits using CottonSNP63K array in upland cotton (Gossypium hirsutum L.). BMC Genom. 2019, 20, 889. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Jiang, Y.; Zhang, W.; Cheng, Y.; Wang, Y.; Ma, X.; Duan, Y.; Xia, L.; Chen, Y.; Wu, N. Construction of a high-density genetic map and mapping of growth-related QTLs in the grass carp (Ctenopharyngodon idellus). BMC Genom. 2020, 21, 313. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, W.; Zhang, J.; Wang, N.; Zhao, Y.; Wang, Y.; Bai, S. Construction of the first high-density genetic linkage map and identification of seed yield-related QTLs and candidate genes in Elymus sibiricus, an important forage grass in Qinghai-Tibet Plateau. BMC Genom. 2019, 20, 861. [Google Scholar] [CrossRef]
- Du, Q.; Yang, X.; Xie, J.; Quan, M.; Xiao, L.; Lu, W.; Tian, J.; Gong, C.; Chen, J.; Li, B. Time-specific and pleiotropic quantitative trait loci coordinately modulate stem growth in Populus. Plant Biotechnol. J. 2019, 17, 608–624. [Google Scholar] [CrossRef] [Green Version]
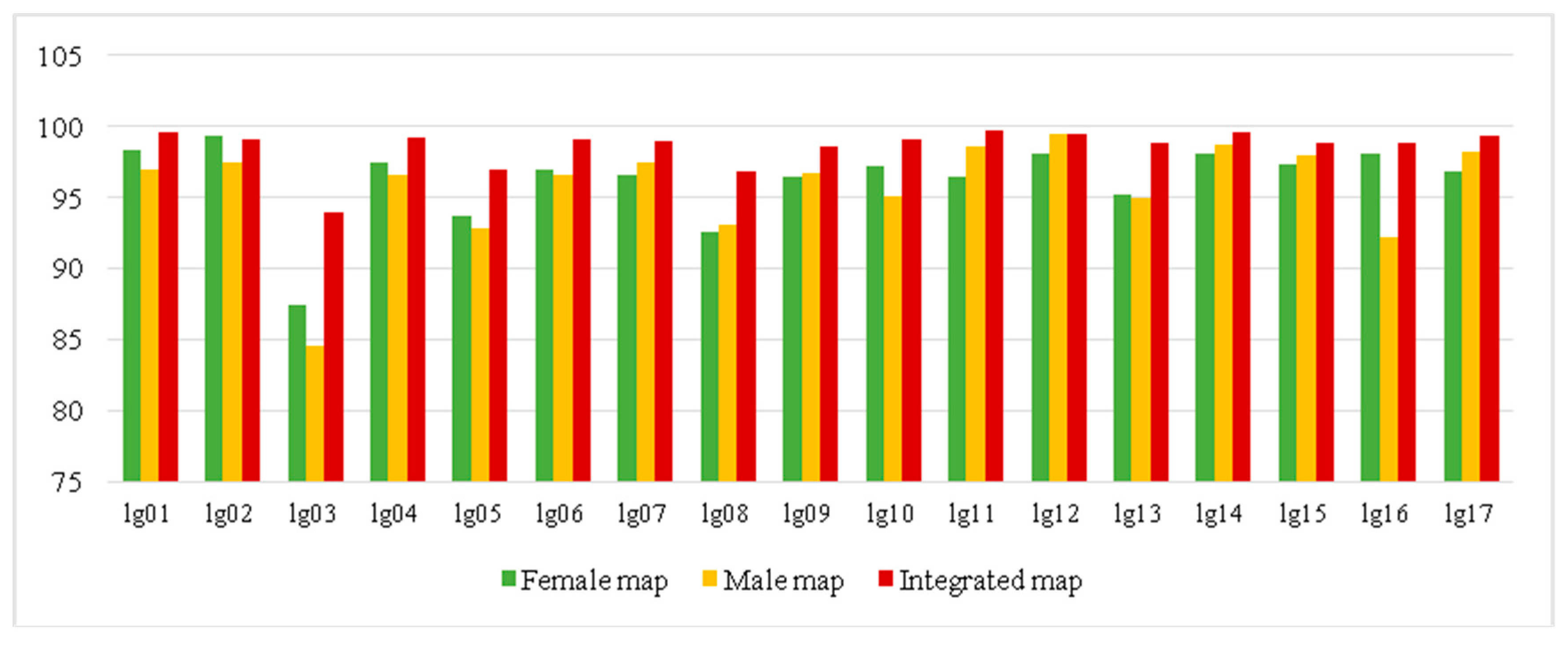
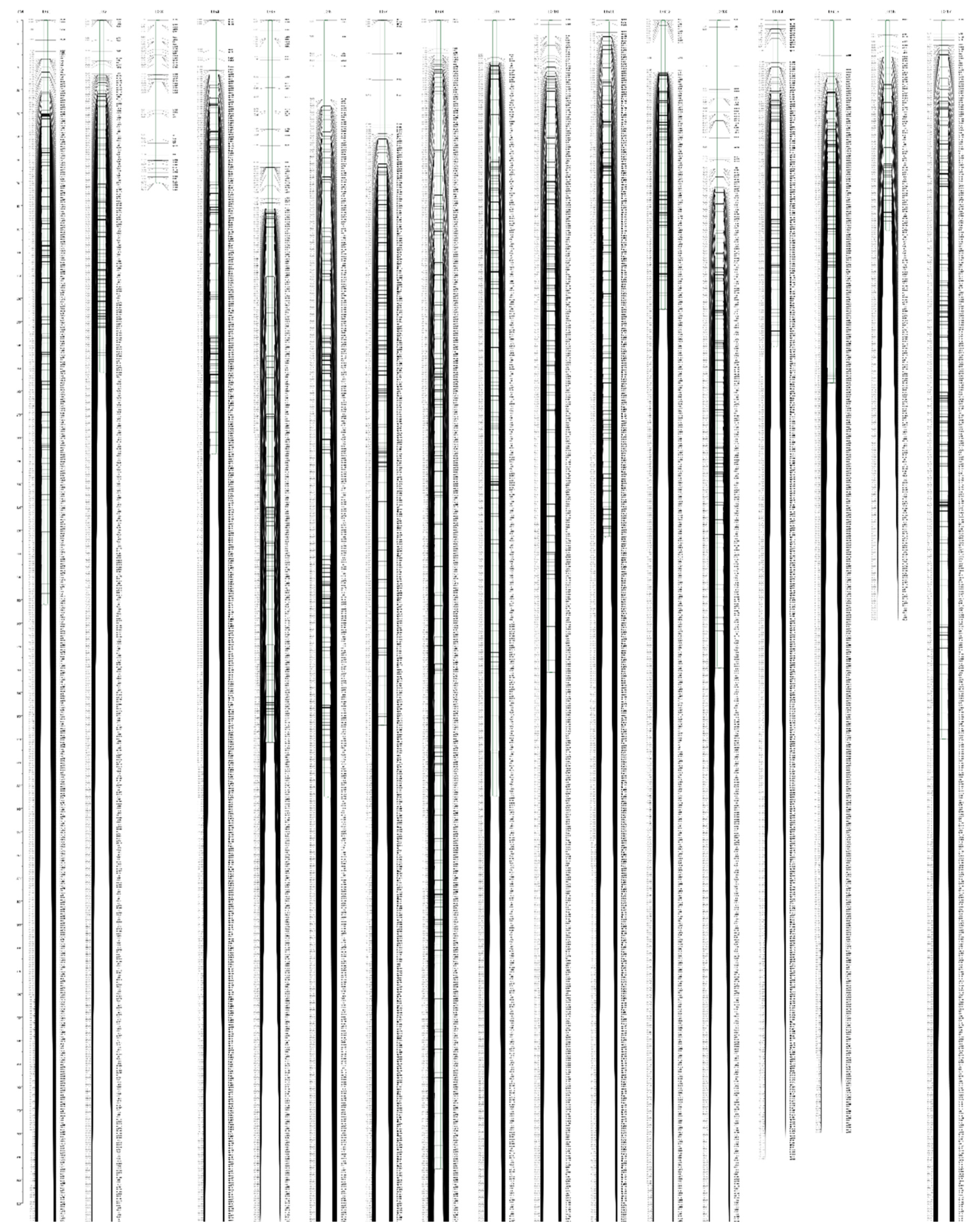

| Linkage Group | Number of Markers | Genetic Length (cM) | Average Marker Interval (cM) | Maximum Gap (cM) | <5 cM |
|---|---|---|---|---|---|
| lg01 | 681 | 252.09 | 0.37 | 8.45 | 678 |
| lg02 | 554 | 152.24 | 0.27 | 8.04 | 548 |
| lg03 | 51 | 70.42 | 1.38 | 12.35 | 47 |
| lg04 | 606 | 187.31 | 0.31 | 12.86 | 600 |
| lg05 | 499 | 311.53 | 0.62 | 10.41 | 483 |
| lg06 | 909 | 335.37 | 0.37 | 14.16 | 900 |
| lg07 | 785 | 304.16 | 0.39 | 14.89 | 776 |
| lg08 | 768 | 495.49 | 0.65 | 12.44 | 743 |
| lg09 | 718 | 334.9 | 0.47 | 18.69 | 707 |
| lg10 | 680 | 281.36 | 0.41 | 10.31 | 673 |
| lg11 | 629 | 223.26 | 0.35 | 5.48 | 626 |
| lg12 | 562 | 124.84 | 0.22 | 12.75 | 558 |
| lg13 | 513 | 279.42 | 0.54 | 15.99 | 506 |
| lg14 | 487 | 141.04 | 0.29 | 5.49 | 484 |
| lg15 | 461 | 156.59 | 0.34 | 15.4 | 455 |
| lg16 | 252 | 90.94 | 0.36 | 7.73 | 248 |
| lg17 | 948 | 310.14 | 0.33 | 11.41 | 941 |
| Average | 594 | 238.3 | 0.45 | / | 587 |
| Total | 10,103 | 4051.11 | / | / | / |
| Group | Position 1 | Locus | Type | LOD | Variance | PVE% | Traits | Scaffold | Position 2 | Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| LG07 | 192.873 | lm3422 | <lmxll> | 2.97 | 83.9875 | 11.8 | NS | Super-Scaffold_31 | 430,546 | EUC26133-RA |
| LG07 | 192.873 | lm3466 | <lmxll> | 2.97 | 83.988 | 11.8 | NS | Super-Scaffold_102 | 2,268,088 | EUC18364-RA |
| LG07 | 106.208 | lm5491 | <lmxll> | 2.95 | 84.0443 | 11.7 | NS | Super-Scaffold_58 | 2,488,749 | EUC19299-RA |
| LG07 | 121.688 | lm5514 | <lmxll> | 2.63 | 85.1833 | 10.5 | NS | Super-Scaffold_58 | 7,131,719 | EUC12085-RA |
| LG07 | 88.094 | lm790 | <lmxll> | 2.63 | 85.2124 | 10.5 | NS | Super-Scaffold_5 | 657,894 | EUC11205-RA |
| LG07 | 89.954 | lm798 | <lmxll> | 2.63 | 85.212 | 10.5 | NS | Super-Scaffold_5 | 812,651 | EUC19303-RA |
| LG07 | 66.34 | lm812 | <lmxll> | 2.95 | 84.0437 | 11.7 | NS | Super-Scaffold_5 | 1,939,567 | EUC12116-RA |
| LG07 | 183.599 | np3236 | <nnxnp> | 2.96 | 84.0324 | 11.7 | NS | Super-Scaffold_31 | 1,620,296 | EUC18384-RA |
| LG07 | 191.476 | np3308 | <nnxnp> | 2.75 | 84.7757 | 11 | NS | Super-Scaffold_31 | 4,766,453 | EUC16568-RA |
| LG07 | 191.014 | np744 | <nnxnp> | 2.97 | 83.9873 | 11.8 | NS | Super-Scaffold_5 | 4,487,860 | EUC18378-RA |
| LG07 | 192.403 | np748 | <nnxnp> | 2.99 | 83.9021 | 11.9 | NS | Super-Scaffold_5 | 4,819,249 | EUC16566-RA |
| LG07 | 107.224 | lm1549 | <lmxll> | 2.97 | 83.9757 | 11.8 | NS | Super-Scaffold_10 | 5,318,207 | NA |
| LG02 | 8.042 | lm972 | <lmxll> | 2.79 | 85.042 | 11.1 | NS | scaffold63068_obj | 7897 | EUC20037-RA |
| LG02 | 8.424 | np3071 | <nnxnp> | 2.76 | 82.424 | 11 | NS | scaffold352_obj | 165,986 | NA |
| LG07 | 188.762 | np3234 | <nnxnp> | 3.7 | 188.762 | 12.6 | NS | Super-Scaffold_31 | 1,609,846 | NA |
| LG07 | 189.163 | np752 | <nnxnp> | 3.82 | 189.163 | 13.4 | NS | Super-Scaffold_5 | 5,878,641 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Wang, L.; Lu, W.; Zhong, J.; Du, H.; Liu, P.; Du, Q.; Du, L.; Qing, J. Construction of SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Growth Traits in Eucommia ulmoides Oliver. Forests 2022, 13, 1479. https://doi.org/10.3390/f13091479
Liu C, Wang L, Lu W, Zhong J, Du H, Liu P, Du Q, Du L, Qing J. Construction of SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Growth Traits in Eucommia ulmoides Oliver. Forests. 2022; 13(9):1479. https://doi.org/10.3390/f13091479
Chicago/Turabian StyleLiu, Chenlu, Lu Wang, Wenjie Lu, Jian Zhong, Hongyan Du, Panfeng Liu, Qingxin Du, Lanying Du, and Jun Qing. 2022. "Construction of SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Growth Traits in Eucommia ulmoides Oliver" Forests 13, no. 9: 1479. https://doi.org/10.3390/f13091479
APA StyleLiu, C., Wang, L., Lu, W., Zhong, J., Du, H., Liu, P., Du, Q., Du, L., & Qing, J. (2022). Construction of SNP-Based High-Density Genetic Map Using Genotyping by Sequencing (GBS) and QTL Analysis of Growth Traits in Eucommia ulmoides Oliver. Forests, 13(9), 1479. https://doi.org/10.3390/f13091479






