Extracellular Enzyme Activity and Stoichiometry Reveal Nutrient Dynamics during Microbially-Mediated Plant Residue Transformation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design
2.3. Measurement
2.4. Statistical Analyses
3. Results
3.1. Variations in Soil Extracellular Enzyme Activities (EEAs) and Extracellular Enzymatic Stoichiometry (EES)
3.1.1. Soil Extracellular Enzyme Activities (EEAs)
3.1.2. Soil Extracellular Enzymatic Stoichiometry (EES)
3.2. Correlation of Soil Extracellular Enzyme Activities with C, N, and P Transformations
3.2.1. Correlation between Soil Extracellular Enzyme Activities and Substrate C
3.2.2. Correlation between Soil Extracellular Enzyme Activities and Substrate N
3.2.3. Correlation between Soil Extracellular Enzyme Activities and Substrate P
3.3. Influential Factors of Soil Extracellular Enzyme Activities and Extracellular Enzymatic Stoichiometry
3.3.1. Influential Factors of Soil Extracellular Enzyme Activities
3.3.2. Influential Factors of Extracellular Enzymatic Stoichiometry
4. Discussion
4.1. Association of Extracellular Enzyme Activities (EEAs) with Environmental Nutrient Variables
4.2. Driving Factors of Soil Enzymatic Stoichiometry (EES)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cotrufo, M.F.; Soong, J.L.; Horton, A.J.; Campbell, E.E.; Haddix, M.L.; Wall, D.H.; Parton, W.J. Formation of soil organic matter via biochemical and physical pathways of litter mass loss. Nat. Geosci. 2015, 8, 776–779. [Google Scholar] [CrossRef]
- Miltner, A.; Bombach, P.; Schmidt-Brücken, B.; Kästner, M. SOM genesis: Microbial biomass as a significant source. Biodegradation 2012, 111, 41–55. [Google Scholar] [CrossRef]
- Sokol, N.W.; Bradford, M.A. Microbial formation of stable soil carbon is more efficient from belowground than aboveground input. Nat. Geosci. 2018, 12, 46–53. [Google Scholar] [CrossRef]
- Clemmensen, K.E.; Bahr, A.; Ovaskainen, O.; Dahlberg, A.; Ekblad, A.; Wallander, H.; Stenlid, J.; Finlay, R.D.; Wardle, D.A.; Lindahl, B.D. Roots and Associated Fungi Drive Long-Term Carbon Sequestration in Boreal Forest. Science 2013, 339, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, S.; Barot, S.; Barré, P.; Bdioui, N.; Mary, B.; Rumpel, C. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 2007, 450, 277–280. [Google Scholar] [CrossRef]
- Berg, B.; Davey, M.P.; De Marco, A.; Emmett, B.; Faituri, M.; Hobbie, S.; Johansson, M.-B.; Liu, C.; McClaugherty, C.; Norell, L.; et al. Factors influencing limit values for pine needle litter decomposition: A synthesis for boreal and temperate pine forest systems. Biogeochemistry 2010, 100, 57–73. [Google Scholar] [CrossRef]
- Chen, L.; Fang, K.; Wei, B.; Qin, S.; Feng, X.; Hu, T.; Ji, C.; Yang, Y. Soil carbon persistence governed by plant input and mineral protection at regional and global scales. Ecol. Lett. 2021, 24, 1018–1028. [Google Scholar] [CrossRef]
- De Marco, A.; Esposito, F.; Berg, B.; Zarrelli, A.; De Santo, A.V. Litter Inhibitory Effects on Soil Microbial Biomass, Activity, and Catabolic Diversity in Two Paired Stands of Robinia pseudoacacia L. and Pinus nigra Arn. Forests 2018, 9, 766. [Google Scholar] [CrossRef] [Green Version]
- Mooshammer, M.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A.A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Schimel, J.P.; Jastrow, J.D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2017, 2, 17105. [Google Scholar] [CrossRef]
- Whalen, E.D.; Grandy, A.S.; Sokol, N.W.; Keiluweit, M.; Ernakovich, J.; Smith, R.G.; Frey, S.D. Clarifying the evidence for microbial- and plant-derived soil organic matter, and the path toward a more quantitative understanding. Glob. Chang. Biol. 2022, 28, 7167–7185. [Google Scholar] [CrossRef] [PubMed]
- Fanin, N.; Moorhead, D.; Bertrand, I. Eco-enzymatic stoichiometry and enzymatic vectors reveal differential C, N, P dynamics in decaying litter along a land-use gradient. Biogeochemistry 2016, 129, 21–36. [Google Scholar] [CrossRef]
- Liang, C.; Kästner, M.; Joergensen, R.G. Microbial necromass on the rise: The growing focus on its role in soil organic matter development. Soil Biol. Biochem. 2020, 150, 108000. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, S.; Shen, H.; Zhao, M.; Xu, L.; Xing, A.; Fang, J. Soil extracellular enzyme activity and stoichiometry in China’s forests. Funct. Ecol. 2020, 34, 1461–1471. [Google Scholar] [CrossRef]
- Bai, X.; Dippold, M.A.; An, S.; Wang, B.; Zhang, H.; Loeppmann, S. Extracellular enzyme activity and stoichiometry: The effect of soil microbial element limitation during leaf litter decomposition. Ecol. Indic. 2020, 121, 107200. [Google Scholar] [CrossRef]
- De Marco, A.; Vittozzi, P.; De Santo, A.V. Elements dynamics, from leaf to stable leaf litter residue and soil, for two functional types of tree planted on volcanic deposits. Plant Soil 2022, 1–14. [Google Scholar] [CrossRef]
- Sokol, N.W.; Sanderman, J.; Bradford, M.A. Pathways of mineral-associated soil organic matter formation: Integrating the role of plant carbon source, chemistry, and point of entry. Glob. Chang. Biol. 2018, 25, 12–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Wang, X.; Zhang, X.; Ju, W.; Duan, C.; Guo, X.; Wang, Y.; Fang, L. Soil moisture mediates microbial carbon and phosphorus metabolism during vegetation succession in a semiarid region. Soil Biol. Biochem. 2020, 147, 107814. [Google Scholar] [CrossRef]
- Agri, U.; Chaudhary, P.; Sharma, A.; Kukreti, B. Physiological response of maize plants and its rhizospheric microbiome under the influence of potential bioinoculants and nanochitosan. Plant Soil 2022, 474, 451–468. [Google Scholar] [CrossRef]
- Kukreti, B.; Sharma, A.; Chaudhary, P.; Agri, U.; Maithani, D. Influence of nanosilicon dioxide along with bioinoculants on Zea mays and its rhizospheric soil. 3 Biotech 2020, 10, 345. [Google Scholar] [CrossRef]
- Kumari, S.; Sharma, A.; Chaudhary, P.; Khati, P. Management of plant vigor and soil health using two agriusable nanocompounds and plant growth promotory rhizobacteria in Fenugreek. 3 Biotech 2020, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Schleuss, P.-M.; Widdig, M.; Heintz-Buschart, A.; Guhr, A.; Martin, S.; Kirkman, K.; Spohn, M. Stoichiometric controls of soil carbon and nitrogen cycling after long-term nitrogen and phosphorus addition in a mesic grassland in South Africa. Soil Biol. Biochem. 2019, 135, 294–303. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Tan, X.; Nie, Y.; Ma, X.; Guo, Z.; Liu, Y.; Tian, H.; Megharaj, M.; Shen, W.; He, W. Soil chemical properties rather than the abundance of active and potentially active microorganisms control soil enzyme kinetics. Sci. Total. Environ. 2021, 770, 144500. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Trivedi, C.; Hu, H.; Anderson, I.; Jeffries, T.; Zhou, J.; Singh, B.K. Microbial regulation of the soil carbon cycle: Evidence from gene–enzyme relationships. ISME J. 2016, 10, 2593–2604. [Google Scholar] [CrossRef] [Green Version]
- Zechmeister-Boltenstern, S.; Keiblinger, K.M.; Mooshammer, M.; Peñuelas, J.; Richter, A.; Sardans, J.; Wanek, W. The application of ecological stoichiometry to plant–microbial–soil organic matter transformations. Ecol. Monogr. 2015, 85, 133–155. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.H.; Gillooly, J.F.; Allen, A.P.; Savage, V.M.; West, G.B. Toward a metabolic theory of ecology. Ecology 2004, 85, 1771–1789. [Google Scholar] [CrossRef]
- Allen, A.P.; Gillooly, J.F. Towards an integration of ecological stoichiometry and the metabolic theory of ecology to better understand nutrient cycling. Ecol. Lett. 2009, 12, 369–384. [Google Scholar] [CrossRef]
- Cui, Y.; Moorhead, D.L.; Guo, X.; Peng, S.; Wang, Y.; Zhang, X.; Fang, L.; Xu, X. Stoichiometric models of microbial metabolic limitation in soil systems. Glob. Ecol. Biogeogr. 2021, 30, 2297–2311. [Google Scholar] [CrossRef]
- Xu, Z.; Yu, G.; Zhang, X.; He, N.; Wang, Q.; Wang, S.; Wang, R.; Zhao, N.; Jia, Y.; Wang, C. Soil enzyme activity and stoichiometry in forest ecosystems along the North-South Transect in eastern China (NSTEC). Soil Biol. Biochem. 2017, 104, 152–163. [Google Scholar] [CrossRef]
- Deng, L.; Peng, C.; Huang, C.; Wang, K.; Liu, Q.; Liu, Y.; Hai, X.; Shangguan, Z. Drivers of soil microbial metabolic limitation changes along a vegetation restoration gradient on the Loess Plateau, China. Geoderma 2019, 353, 188–200. [Google Scholar] [CrossRef]
- Sokol, N.W.; Kuebbing, S.E.; Karlsen-Ayala, E.; Bradford, M. Evidence for the primacy of living root inputs, not root or shoot litter, in forming soil organic carbon. New Phytol. 2018, 221, 233–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angst, G.; Mueller, K.E.; Nierop, K.G.; Simpson, M.J. Plant- or microbial-derived? A review on the molecular composition of stabilized soil organic matter. Soil Biol. Biochem. 2021, 156, 108189. [Google Scholar] [CrossRef]
- Lehmann, J.; Kleber, M. The contentious nature of soil organic matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Kandeler, E. Xylanase, invertase and protease at the soil–litter interface of a loamy sand. Soil Biol. Biochem. 1999, 31, 1171–1179. [Google Scholar] [CrossRef]
- Ai, C.; Liang, G.; Sun, J.; Wang, X.; Zhou, W. Responses of extracellular enzyme activities and microbial community in both the rhizosphere and bulk soil to long-term fertilization practices in a fluvo-aquic soil. Geoderma 2012, 173–174, 330–338. [Google Scholar] [CrossRef]
- Prescott, C.E.; Vesterdal, L. Decomposition and transformations along the continuum from litter to soil organic matter in forest soils. For. Ecol. Manag. 2021, 498, 119522. [Google Scholar] [CrossRef]
- Xue, Z.; Liu, C.; Zhou, Z.; Wanek, W. Extracellular enzyme stoichiometry reflects the metabolic C-and P-limitations along a grassland succession on the Loess Plateau in China. Appl. Soil Ecol. 2022, 179, 104594. [Google Scholar] [CrossRef]
- Xue, Z. The Decomposition Characteristics of Typical Plant Litters on Grassland Ecosystem on Loess Hilly-Gully Region. Ph.D. Thesis, Northwest A&F University, Yangling, China, 2015. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis: Part 3 Chemical Methods; SSSA Book Series No. 5, SSSA and ASA; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Soltanpour, P.N., Tabatabai, M.A., Johnston, C.T., Sumner, M.E., Eds.; American Society of Agronomy: Madison, WI, USA, 1996; pp. 961–1010. [Google Scholar]
- Bremner, J.M.; Mulvaney, C.S. Nitrogen-Total. In Methods of Soil Analysis: Part 2 Chemical and Microbiological Properties; Bremner, J.M., Mulvaney, C.S., Eds.; American Society of Agronomy: Madison, WI, USA, 1983; pp. 595–624. [Google Scholar]
- Olsen, S.R.; Sommers, L. Methods of Soil Analysis, Part 2, Chemical and Microbial Properties. In Agronomy Monograph; Page, A.L., Miller, R.H., Keeney, D.R., Olsen, S.R., Sommers, L., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; Volume 2, pp. 403–430. [Google Scholar]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Brookes, P.; Landman, A.; Pruden, G.; Jenkinson, D. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Wu, J.; Joergensen, R.G.; Pommerening, B.; Chaussod, R.; Brookes, P.C. Measurement of soil microbial biomass C by fumigation-extraction—An automated procedure. Soil Biol. Biochem. 1990, 22, 1167–1169. [Google Scholar] [CrossRef]
- Li, J.; Cooper, J.M.; Lin, Z.; Li, Y.; Yang, X.; Zhao, B. Soil microbial community structure and function are significantly affected by long-term organic and mineral fertilization regimes in the North China Plain. Appl. Soil Ecol. 2015, 96, 75–87. [Google Scholar] [CrossRef]
- Wang, B.; Liang, C.; Yao, H.; Yang, E.; An, S. The accumulation of microbial necromass carbon from litter to mineral soil and its contribution to soil organic carbon sequestration. Catena 2021, 207, 105622. [Google Scholar] [CrossRef]
- Saiya-Cork, K.; Sinsabaugh, R.; Zak, D. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 2002, 34, 1309–1315. [Google Scholar] [CrossRef]
- Cui, Y.; Fang, L.; Deng, L.; Guo, X.; Han, F.; Ju, W.; Wang, X.; Chen, H.; Tan, W.; Zhang, X. Patterns of soil microbial nutrient limitations and their roles in the variation of soil organic carbon across a precipitation gradient in an arid and semi-arid region. Sci. Total. Environ. 2018, 658, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, D.L.; Sinsabaugh, R.L.; Hill, B.H.; Weintraub, M.N. Vector analysis of ecoenzyme activities reveal constraints on coupled C, N and P dynamics. Soil Biol. Biochem. 2016, 93, 1–7. [Google Scholar] [CrossRef]
- Soong, J.L.; Fuchslueger, L.; Marañon-Jimenez, S.; Torn, M.S.; Janssens, I.A.; Penuelas, J.; Richter, A. Microbial carbon limitation: The need for integrating microorganisms into our understanding of ecosystem carbon cycling. Glob. Chang. Biol. 2019, 26, 1953–1961. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Zheng, H.; Vesterdal, L.; Schmidt, I.K.; Rousk, J. Ecoenzymatic stoichiometry can reflect microbial resource limitation, substrate quality, or both in forest soils. Soil Biol. Biochem. 2022, 167, 108613. [Google Scholar] [CrossRef]
- Chomel, M.; Guittonny-Larchevêque, M.; Fernandez, C.; Gallet, C.; DesRochers, A.; Paré, D.; Jackson, B.G.; Baldy, V. Plant secondary metabolites: A key driver of litter decomposition and soil nutrient cycling. J. Ecol. 2016, 104, 1527–1541. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, X.; Liu, L.; Li, T.; Dou, Y.; Qiao, J.; Wang, Y.; An, S.; Chang, S.X. Nitrogen fertilization weakens the linkage between soil carbon and microbial diversity: A global meta-analysis. Glob. Chang. Biol. 2022, 28, 6446–6461. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, T.; Damris, M.; Kuzyakov, Y. Losses of soil carbon by converting tropical forest to plantations: Erosion and decomposition estimated by δ13C. Glob. Chang. Biol. 2015, 21, 3548–3560. [Google Scholar] [CrossRef] [PubMed]
- Cotrufo, M.F.; Lavallee, J.M. Soil organic matter formation, persistence, and functioning: A synthesis of current understanding to inform its conservation and regeneration. Adv. Agron. 2022, 172, 1–66. [Google Scholar]
- Zhang, X.; Jia, J.; Chen, L.; Chu, H.; He, J.-S.; Zhang, Y.; Feng, X. Aridity and NPP constrain contribution of microbial necromass to soil organic carbon in the Qinghai-Tibet alpine grasslands. Soil Biol. Biochem. 2021, 156, 108213. [Google Scholar] [CrossRef]
- Sayer, E.J.; Wright, S.J.; Tanner, E.V.J.; Yavitt, J.B.; Harms, K.E.; Powers, J.S.; Kaspari, M.; Garcia, M.N.; Turner, B. Variable Responses of Lowland Tropical Forest Nutrient Status to Fertilization and Litter Manipulation. Ecosystems 2012, 15, 387–400. [Google Scholar] [CrossRef]
- Allison, S.D.; Vitousek, P.M. Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol. Biochem. 2005, 37, 937–944. [Google Scholar] [CrossRef]
- Elser, J.J. Phosphorus: A limiting nutrient for humanity? Curr. Opin. Biotechnol. 2012, 23, 833–838. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Shah, J.J.F. Ecoenzymatic Stoichiometry and Ecological Theory. Annu. Rev. Ecol. Evol. Syst. 2012, 43, 313–343. [Google Scholar] [CrossRef] [Green Version]
- Mori, T.; Aoyagi, R.; Kitayama, K.; Mo, J. Does the ratio of β-1,4-glucosidase to β-1,4-N-acetylglucosaminidase indicate the relative resource allocation of soil microbes to C and N acquisition? Soil Biol. Biochem. 2021, 160, 108363. [Google Scholar] [CrossRef]
- Wang, B.; Huang, Y.; Li, N.; Yao, H.; Yang, E.; Soromotin, A.V.; Kuzyakov, Y.; Cheptsov, V.; Yang, Y.; An, S. Initial soil formation by biocrusts: Nitrogen demand and clay protection control microbial necromass accrual and recycling. Soil Biol. Biochem. 2022, 167, 108607. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, C.; Wang, Y.; Cheng, H.; An, S.; Chang, S.X. Soil extracellular enzyme stoichiometry reflects the shift from P- to N-limitation of microorganisms with grassland restoration. Soil Biol. Biochem. 2020, 149, 107928. [Google Scholar] [CrossRef]
- Heuck, C.; Weig, A.; Spohn, M. Soil microbial biomass C:N:P stoichiometry and microbial use of organic phosphorus. Soil Biol. Biochem. 2015, 85, 119–129. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.; Hai, X.; Shangguan, Z.; Deng, L. Dynamics of soil microbial C:N:P stoichiometry and its driving mechanisms following natural vegetation restoration after farmland abandonment. Sci. Total. Environ. 2019, 693, 133613. [Google Scholar] [CrossRef] [PubMed]
- Spohn, M. Element cycling as driven by stoichiometric homeostasis of soil microorganisms. Basic Appl. Ecol. 2016, 17, 471–478. [Google Scholar] [CrossRef]
- Fabian, J.; Zlatanović, S.; Mutz, M.; Premke, K. Fungal–bacterial dynamics and their contribution to terrigenous carbon turnover in relation to organic matter quality. ISME J. 2016, 11, 415–425. [Google Scholar] [CrossRef]
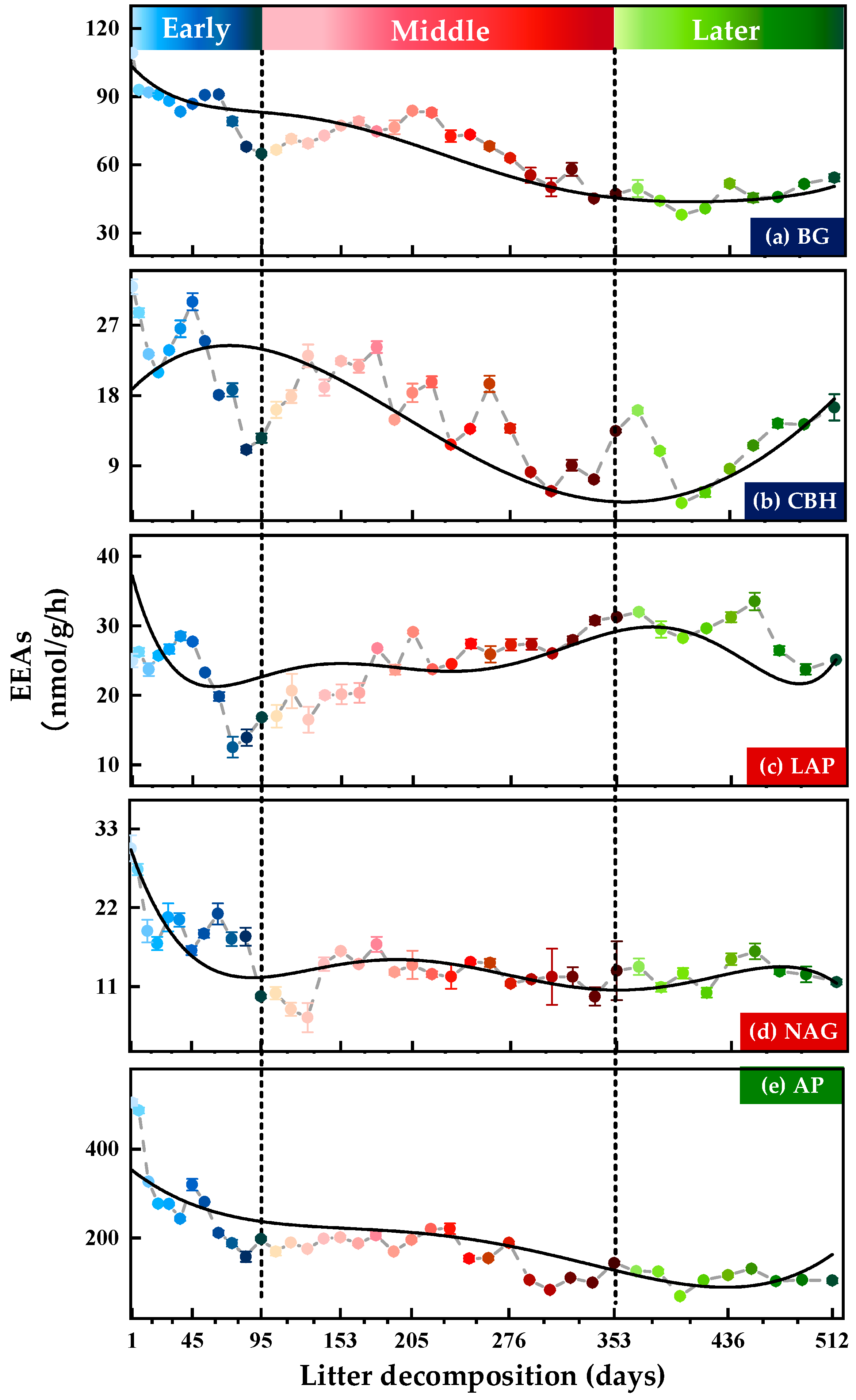
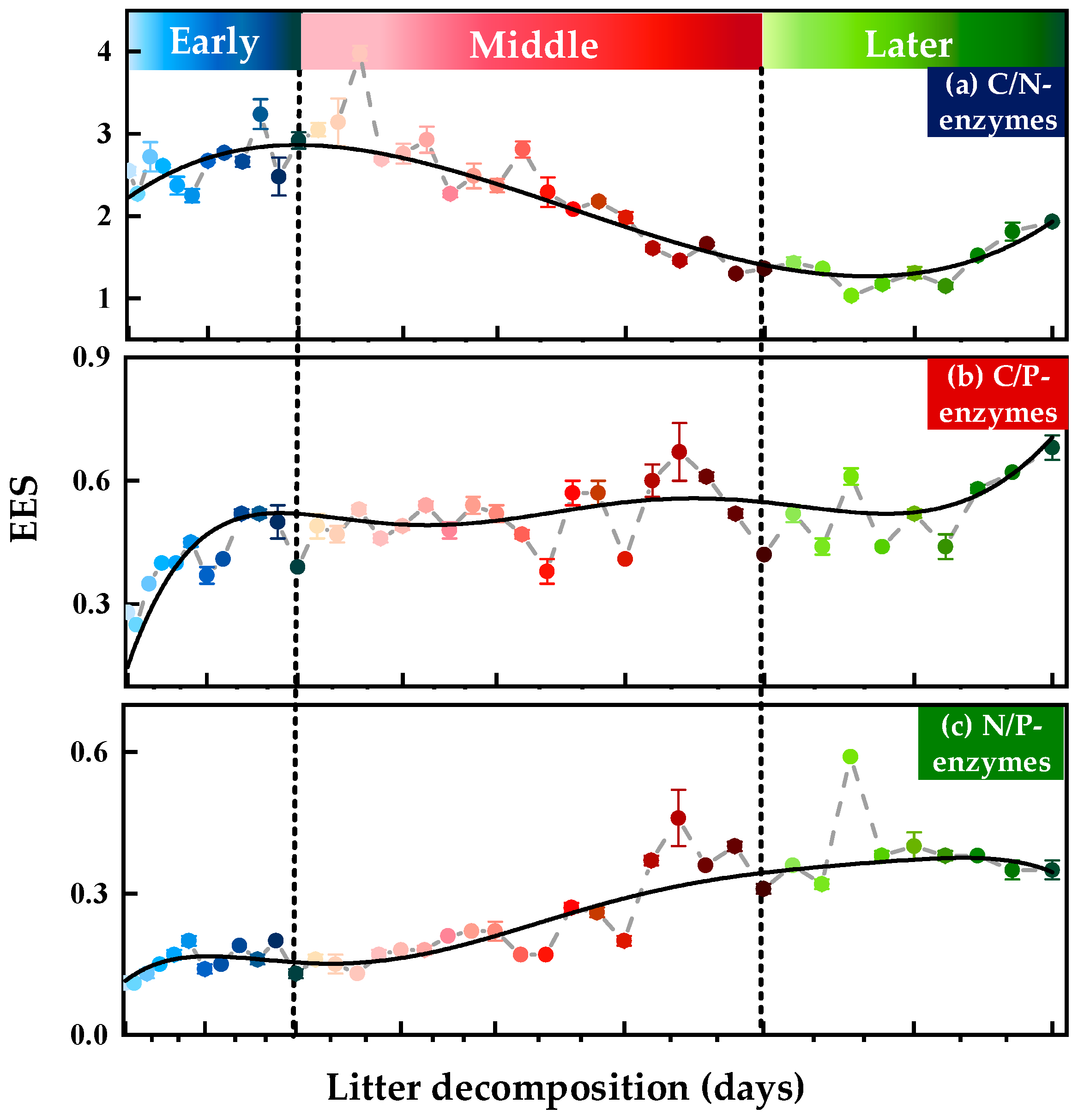


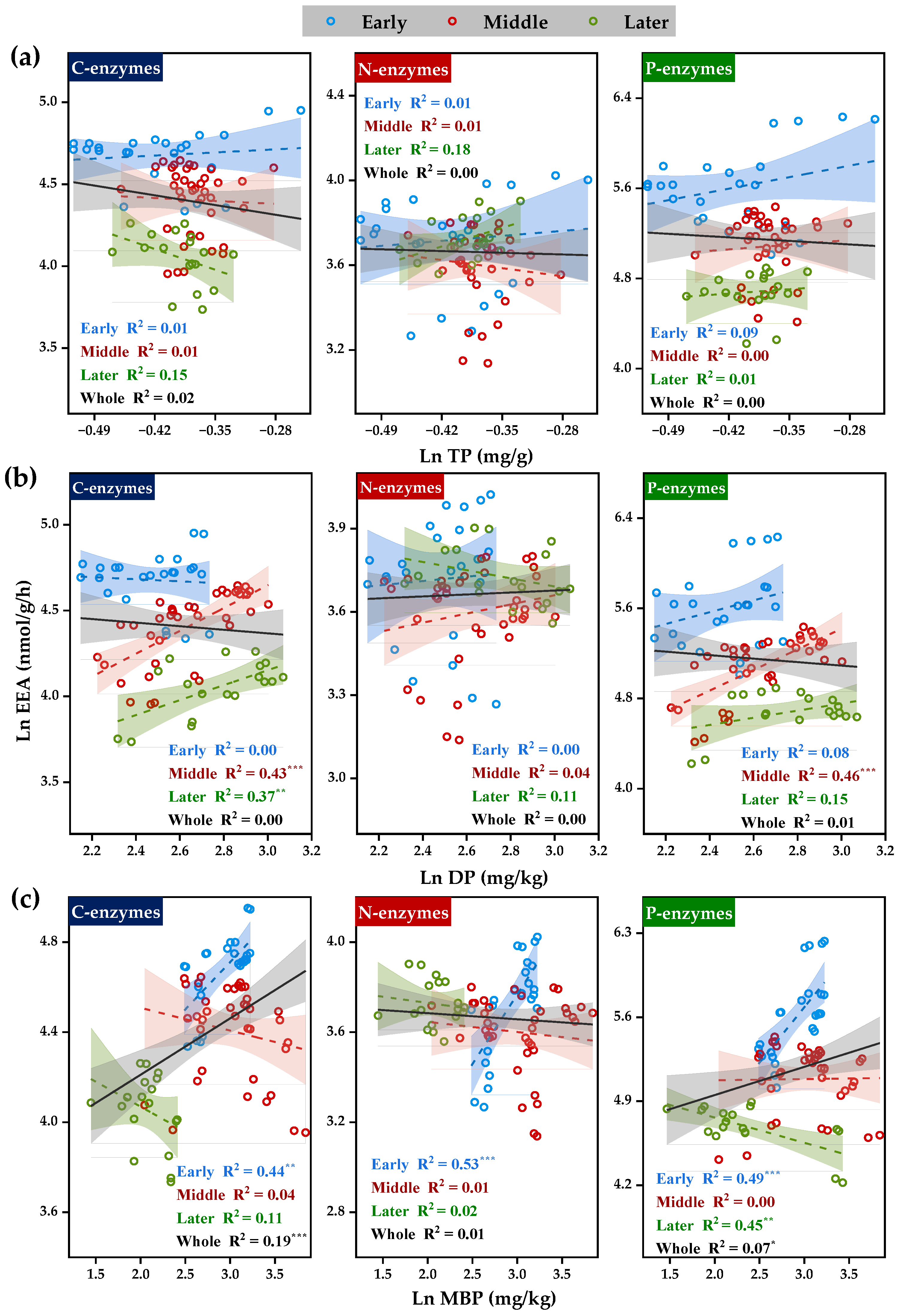

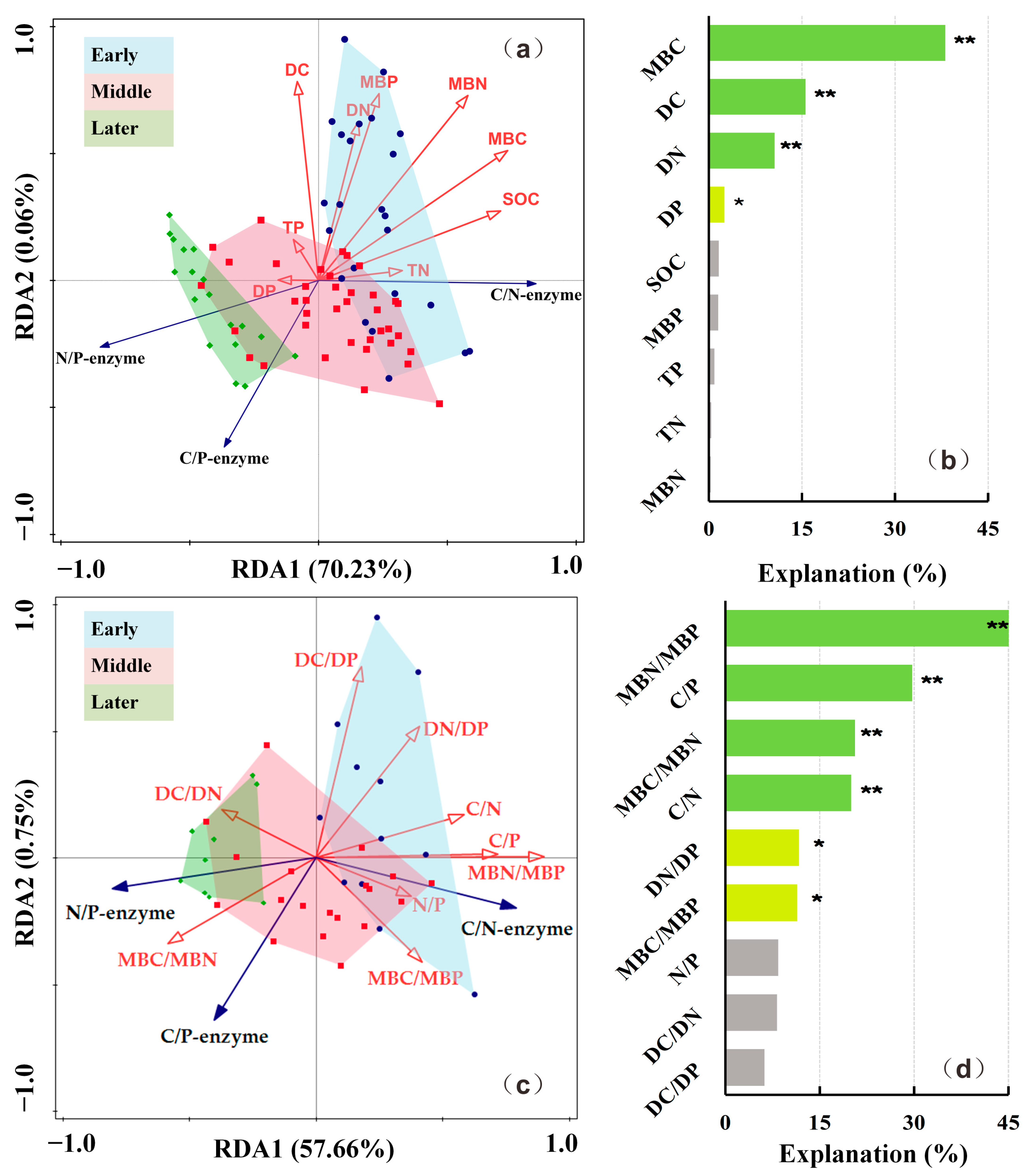
| Correlation Coefficients | BG | CBH | LAP | NAG | AP | |
|---|---|---|---|---|---|---|
| Total C, N, P | SOC | 0.587 ** | 0.526 ** | −0.543 ** | 0.438 ** | 0.548 ** |
| TN | 0.486 ** | 0.444 ** | −0.199 | 0.389 * | 0.402 * | |
| TP | −0.001 | −0.026 | 0.033 | 0.125 | 0.220 | |
| C/N | 0.352 * | 0.320 * | −0.478 ** | 0.248 | 0.364 * | |
| C/P | 0.543 ** | 0.491 ** | −0.499 ** | 0.359 * | 0.422 ** | |
| N/P | 0.395 * | 0.373 * | −0.162 | 0.254 | 0.210 | |
| Microbial biomass C, N, P | MBC | 0.892 ** | 0.731 ** | −0.277 | 0.624 ** | 0.854 ** |
| MBN | 0.790 ** | 0.650 ** | −0.285 | 0.727 ** | 0.850 ** | |
| MBP | 0.538 ** | 0.511 ** | 0.006 | 0.655 ** | 0.682 ** | |
| MBC/MBN | −0.501 ** | −0.313 * | 0.321 * | −0.364 * | −0.464 ** | |
| MBC/MBP | 0.295 | 0.143 | −0.229 | −0.116 | 0.043 | |
| MBN/MBP | 0.700 ** | 0.430 ** | −0.532 ** | 0.288 | 0.511 ** | |
| Dissolved C, N, P | DC | 0.334 * | 0.414 ** | 0.249 | 0.688 ** | 0.624 ** |
| DN | 0.382 * | 0.428 ** | −0.101 | 0.662 ** | 0.617 ** | |
| DP | −0.161 | −0.046 | 0.176 | −0.099 | −0.134 | |
| DC/DN | −0.072 | −0.075 | 0.536 ** | 0.013 | −0.030 | |
| DC/DP | 0.418 ** | 0.445 ** | 0.174 | 0.695 ** | 0.661 ** | |
| DN/DP | 0.459 ** | 0.438 ** | −0.198 | 0.637 ** | 0.616 ** | |
| Correlation Coefficients | C/N-Enzyme | C/P-Enzyme | N/P-Enzyme | |
|---|---|---|---|---|
| Total C, N, P | SOC | 0.598 *** | −0.493 *** | −0.604 *** |
| TN | 0.280 * | −0.147 | −0.282 * | |
| TP | −0.090 | −0.060 | 0.062 | |
| C/N | 0.466 ** | −0.357 * | −0.453 ** | |
| C/P | 0.583 ** | −0.328 * | −0.540 ** | |
| N/P | 0.312 | −0.083 | −0.276 | |
| Microbial biomass C, N, P | MBC | 0.695 *** | −0.502 *** | −0.824 *** |
| MBN | 0.606 *** | −0.647 *** | −0.728 *** | |
| MBP | 0.156 | −0.515 *** | −0.300 * | |
| MBC/MBN | −0.418 ** | 0.470 ** | 0.458 ** | |
| MBC/MBP | 0.363 * | 0.081 | −0.339 * | |
| MBN/MBP | 0.685 ** | −0.378 * | −0.717 ** | |
| Dissolved C, N, P | DC | −0.156 | −0.418 * | −0.046 |
| DN | 0.088 | −0.409 ** | −0.201 | |
| DP | −0.126 | 0.051 | 0.039 | |
| DC/DN | −0.378 * | 0.042 | 0.265 | |
| DC/DP | −0.024 | −0.505 ** | −0.155 | |
| DN/DP | 0.227 | −0.461 ** | −0.317 * | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Ma, J.; Qu, T.; Xue, Z.; Li, X.; Chen, Q.; Wang, N.; Zhou, Z.; An, S. Extracellular Enzyme Activity and Stoichiometry Reveal Nutrient Dynamics during Microbially-Mediated Plant Residue Transformation. Forests 2023, 14, 34. https://doi.org/10.3390/f14010034
Liu C, Ma J, Qu T, Xue Z, Li X, Chen Q, Wang N, Zhou Z, An S. Extracellular Enzyme Activity and Stoichiometry Reveal Nutrient Dynamics during Microbially-Mediated Plant Residue Transformation. Forests. 2023; 14(1):34. https://doi.org/10.3390/f14010034
Chicago/Turabian StyleLiu, Chunhui, Jingyi Ma, Tingting Qu, Zhijing Xue, Xiaoyun Li, Qin Chen, Ning Wang, Zhengchao Zhou, and Shaoshan An. 2023. "Extracellular Enzyme Activity and Stoichiometry Reveal Nutrient Dynamics during Microbially-Mediated Plant Residue Transformation" Forests 14, no. 1: 34. https://doi.org/10.3390/f14010034
APA StyleLiu, C., Ma, J., Qu, T., Xue, Z., Li, X., Chen, Q., Wang, N., Zhou, Z., & An, S. (2023). Extracellular Enzyme Activity and Stoichiometry Reveal Nutrient Dynamics during Microbially-Mediated Plant Residue Transformation. Forests, 14(1), 34. https://doi.org/10.3390/f14010034








