Transcriptome Analysis Reveals the Molecular Mechanisms Associated with Flower Color Formation in Camellia japonica ‘Joy Kendrick’
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. Determination of Anthocyanin Content
2.3. Transcriptome Analysis
2.4. De Novo Assembly and Functional Annotation of Unigenes
2.5. Differential Expression Analysis
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Statistical Analysis
3. Results
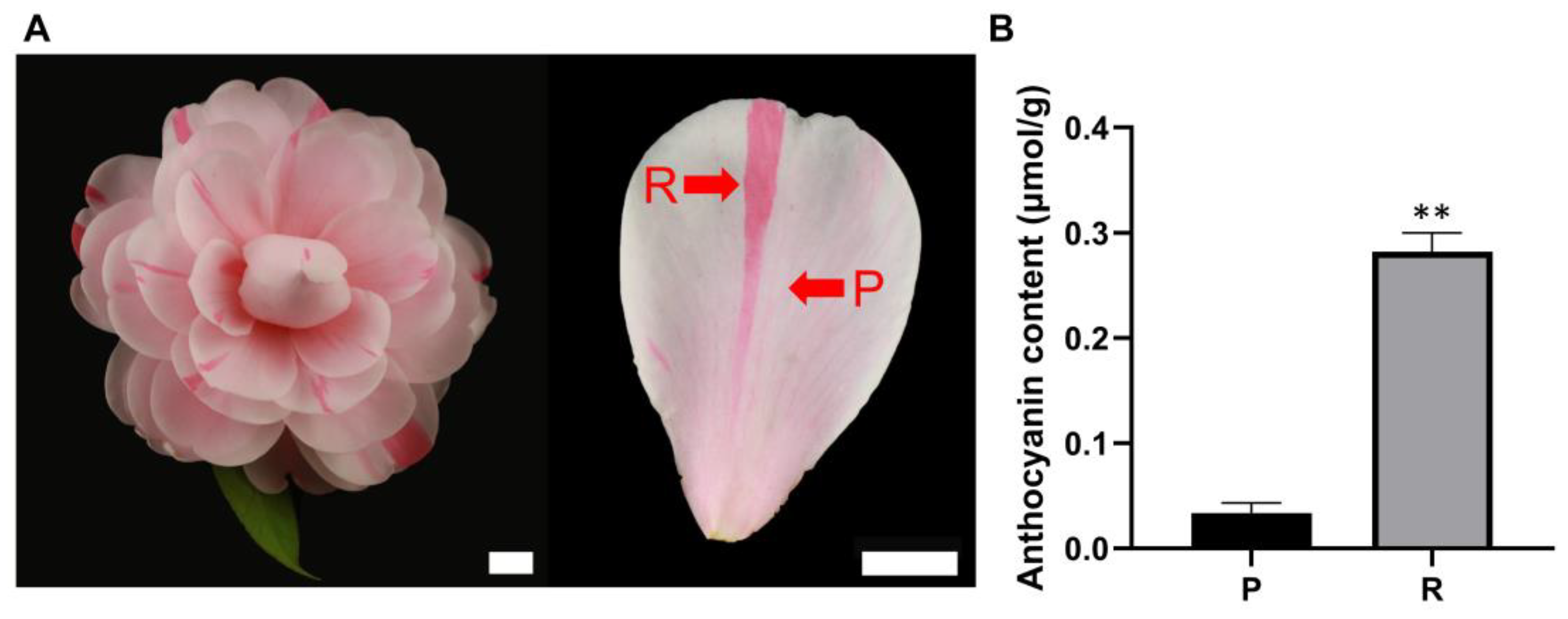
3.1. Phenotypic Characteristics and Anthocyanin Analysis of ‘Joy Kendrick’
3.2. Transcriptome Data Overview
3.3. Identification and Enrichment of DEGs
3.4. Gene Set Enrichment Analysis
3.5. Identification of Candidate Genes Involved in Anthocyanin Biosynthesis
3.6. Plant Hormone Signal Transduction
3.7. Identification of Transcription Factors Regulating Color Formation in ‘Joy Kendrick’
3.8. Validation of Candidate Genes by Quantitative Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sasaki, N.; Nakayama, T. Achievements and Perspectives in Biochemistry Concerning Anthocyanin Modification for Blue Flower Coloration. Plant Cell Physiol. 2015, 56, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Mahmood, K.; Rothstein, S.J. ROS induces anthocyanin production via late biosynthetic genes and anthocyanin deficiency confers the hypersensitivity to ROS-generating stresses in Arabidopsis. Plant Cell Physiol. 2017, 58, 1364–1377. [Google Scholar] [CrossRef] [Green Version]
- Moustaka, J.; Panteris, E.; Adamakis, I.-D.S.; Tanou, G.; Giannakoula, A.; Eleftheriou, E.P.; Moustakas, M. High anthocyanin accumulation in poinsettia leaves is accompanied by thylakoid membrane unstacking, acting as a photoprotective mechanism, to prevent ROS formation. Environ. Exp. Bot. 2018, 154, 44–55. [Google Scholar] [CrossRef]
- Zhu, Y.; Ling, W.; Guo, H.; Song, F.; Ye, Q.; Zou, T.; Li, D.; Zhang, Y.; Li, G.; Xiao, Y. Anti-inflammatory effect of purified dietary anthocyanin in adults with hypercholesterolemia: A randomized controlled trial. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 843–849. [Google Scholar] [CrossRef]
- Verediano, T.A.; Stampini Duarte Martino, H.; Dias Paes, M.C.; Tako, E. Effects of anthocyanin on intestinal health: A systematic review. Nutrients 2021, 13, 1331. [Google Scholar] [CrossRef]
- Sonia, D.P.-T.; Teresa, S.-B.M. Anthocyanins: From plant to health. Phytochem. Rev. 2008, 7, 281–299. [Google Scholar]
- Bowen-Forbes, C.S.; Zhang, Y.; Nair, M.G. Anthocyanin content, antioxidant, anti-inflammatory and anticancer properties of blackberry and raspberry fruits. J. Food Compos. Anal. 2010, 23, 554–560. [Google Scholar] [CrossRef]
- Panchal, S.K.; John, O.D.; Mathai, M.L.; Brown, L. Anthocyanins in Chronic Diseases: The Power of Purple. Nutrients 2022, 14, 2161. [Google Scholar] [CrossRef]
- Roberto, M.; Antonio, F.; Luciana, M.; Paula, S. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 3809. [Google Scholar]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Shen, Y.; Sun, T.; Pan, Q.; Anupol, N.; Chen, H.; Shi, J.; Liu, F.; Deqiang, D.; Wang, C.; Zhao, J. Rr MYB 5-and Rr MYB 10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 2019, 17, 2078–2095. [Google Scholar] [CrossRef] [Green Version]
- Mao, W.; Han, Y.; Chen, Y.; Sun, M.; Feng, Q.; Li, L.; Liu, L.; Zhang, K.; Wei, L.; Han, Z. Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of Chalcone Synthase 1. Plant Cell 2022, 34, 1226–1249. [Google Scholar] [CrossRef] [PubMed]
- Karppinen, K.; Lafferty, D.J.; Albert, N.W.; Mikkola, N.; McGhie, T.; Allan, A.C.; Afzal, B.M.; Häggman, H.; Espley, R.V.; Jaakola, L. MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 2021, 232, 1350–1367. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Sharma, N.; Kapoor, P.; Chunduri, V.; Pandey, A.K.; Garg, M. Spotlight on the overlapping routes and partners for anthocyanin transport in plants. Physiol. Plant. 2021, 171, 868–881. [Google Scholar] [CrossRef]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef]
- Li, Q.; Wang, J.; Sun, H.-Y.; Shang, X. Flower color patterning in pansy (Viola × wittrockiana Gams.) is caused by the differential expression of three genes from the anthocyanin pathway in acyanic and cyanic flower areas. Plant Physiol. Biochem. 2014, 84, 134–141. [Google Scholar] [CrossRef]
- Gu, Z.; Zhu, J.; Hao, Q.; Yuan, Y.-W.; Duan, Y.-W.; Men, S.; Wang, Q.; Hou, Q.; Liu, Z.-A.; Shu, Q. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef]
- Shang, Y.; Venail, J.; Mackay, S.; Bailey, P.C.; Schwinn, K.E.; Jameson, P.E.; Martin, C.R.; Davies, K.M. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011, 189, 602–615. [Google Scholar] [CrossRef]
- Simonds, V.; Plowright, C. How do bumblebees first find flowers? Unlearned approach responses and habituation. Anim. Behav. 2004, 67, 379–386. [Google Scholar] [CrossRef]
- Heuschen, B.; Gumbert, A.; Lunau, K. A generalised mimicry system involving angiosperm flower colour, pollen and bumblebees’ innate colour preferences. Plant Syst. Evol. 2005, 252, 121–137. [Google Scholar] [CrossRef]
- Hieber, A.D.; Mudalige-Jayawickrama, R.G.; Kuehnle, A.R. Color genes in the orchid Oncidium Gower Ramsey: Identification, expression, and potential genetic instability in an interspecific cross. Planta 2006, 223, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Shu, Q.; Liu, Z.A.; Li, C.; Zhang, J.; Wei, X.; Tian, D. Comparison of anthocyanins in non-blotches and blotches of the petals of Xibei tree peony. Sci. Hortic. 2007, 114, 104–111. [Google Scholar] [CrossRef]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef] [Green Version]
- Clouse, S.D. The molecular intersection of brassinosteroid-regulated growth and flowering in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 7345–7346. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Liu, Q.; Xi, B.; Dai, H. Study on the regulation of anthocyanin biosynthesis by exogenous abscisic acid in grapevine. Sci. Hortic. 2019, 250, 294–301. [Google Scholar] [CrossRef]
- Gao, H.-N.; Jiang, H.; Cui, J.-Y.; You, C.-X.; Li, Y.-Y. The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 2021, 312, 111024. [Google Scholar] [CrossRef]
- Wang, S.; Li, L.-X.; Fang, Y.; Li, D.; Mao, Z.; Zhu, Z.; Chen, X.-S.; Feng, S.-Q. MdERF1B–MdMYC2 module integrates ethylene and jasmonic acid to regulate the biosynthesis of anthocyanin in apple. Hortic. Res. 2022, 9, uhac142. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYB transcription factor MYB 6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Kundan, M.; Gani, U.; Fayaz, M.; Angmo, T.; Kesari, R.; Rahul, V.P.; Gairola, S.; Misra, P. Two R2R3-MYB transcription factors, CsMYB33 and CsMYB78 are involved in the regulation of anthocyanin biosynthesis in Cannabis sativa L. Ind. Crops Prod. 2022, 188, 115546. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–b HLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.W.; Sagawa, J.M.; Frost, L.; Vela, J.P.; Bradshaw, H.D., Jr. Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus). New Phytol. 2014, 204, 1013–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.M.; Rudall, P.J.; Ellis, A.G.; Savolainen, V.; Glover, B.J. Development of a complex floral trait: The pollinator-attracting petal spots of the beetle daisy, Gorteria diffusa (Asteraceae). Am. J. Bot. 2009, 96, 2184–2196. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, Y.; Ya, H.; Xu, S.; Han, J. Transcriptome sequencing of purple petal spot region in tree peony reveals differentially expressed anthocyanin structural genes. Front. Plant Sci. 2015, 6, 964. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Fan, M.; Zhang, Y.; Yang, M.; Wu, S.; Yin, H.; Li, J.; Li, X. Transcriptomic and Chemical Analyses Reveal the Hub Regulators of Flower Color Variation from Camellia japonica Bud Sport. Horticulturae 2022, 8, 129. [Google Scholar] [CrossRef]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Yang, X.; Tu, Y.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S. Unraveling the regulatory mechanism of color diversity in camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 2021, 12, 1119. [Google Scholar] [CrossRef]
- Xing, Y.; Wang, K.; Huang, C.; Huang, J.; Zhao, Y.; Si, X.; Li, Y. Global Transcriptome Analysis Revealed the Molecular Regulation Mechanism of Pigment and Reactive Oxygen Species Metabolism During the Stigma Development of Carya cathayensis. Front. Plant Sci. 2022, 13, 881394. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chen, W.; Xiang, W.; Wang, D.; Xue, B.; Liu, X.; Xing, L.; Wu, D.; Wang, S.; Guo, Q. Integrated metabolic profiling and transcriptome analysis of pigment accumulation in Lonicera japonica flower petals during colour-transition. BMC Plant Biol. 2021, 21, 98. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.-Y.; Yeh, K.-W. Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol. 2008, 66, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, Q.-Y.; Feng, Z.-H.; Wang, B.; Zhang, Y.-F.; Yang, Q. Increased accumulation of anthocyanins in transgenic potato tubers by overexpressing the 3GT gene. Plant Biotechnol. Rep. 2012, 6, 69–75. [Google Scholar] [CrossRef]
- Sun, W.; Liang, L.; Meng, X.; Li, Y.; Gao, F.; Liu, X.; Wang, S.; Gao, X.; Wang, L. Biochemical and molecular characterization of a flavonoid 3-O-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in Freesia hybrida. Front. Plant Sci. 2016, 7, 410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Wei, G.; Zhou, H.; Gu, C.; Vimolmangkang, S.; Liao, L.; Han, Y. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 2014, 166, 1044–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [Green Version]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Huang, J.-R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 2012, 5, 387–400. [Google Scholar] [CrossRef]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Larsen, E.; Alfenito, M.; Briggs, W.; Walbot, V. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Rep. 2003, 21, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef] [PubMed]
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.-Y.; Dean, J.V. Transport of anthocyanins and other flavonoids by the Arabidopsis ATP-binding cassette transporter AtABCC2. Sci. Rep. 2019, 9, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Zhao, J.; Qin, B.; Yin, Y.; An, W.; Mu, Z.; Cao, Y. ABA mediates development-dependent anthocyanin biosynthesis and fruit coloration in Lycium plants. BMC Plant Biol. 2019, 19, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef]
- Ji, X.-H.; Zhang, R.; Wang, N.; Yang, L.; Chen, X.-S. Transcriptome profiling reveals auxin suppressed anthocyanin biosynthesis in red-fleshed apple callus (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Cult. (PCTOC) 2015, 123, 389–404. [Google Scholar] [CrossRef]
- Moro, L.; Hassimotto, N.M.A.; Purgatto, E. Postharvest auxin and methyl jasmonate effect on anthocyanin biosynthesis in red raspberry (Rubus idaeus L.). J. Plant Growth Regul. 2017, 36, 773–782. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-C.; Wang, N.; Xu, H.-F.; Jiang, S.-H.; Fang, H.-C.; Su, M.-Y.; Zhang, Z.-Y.; Zhang, T.-L.; Chen, X.-S. Auxin regulates anthocyanin biosynthesis through the Aux/IAA–ARF signaling pathway in apple. Hortic. Res. 2018, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-K.; Han, P.-L.; Zhao, Y.-W.; Yu, J.-Q.; You, C.-X.; Hu, D.-G.; Hao, Y.-J. Genome-wide analysis of auxin response factor (ARF) genes and functional identification of MdARF2 reveals the involvement in the regulation of anthocyanin accumulation in apple. N. Z. J. Crop Hortic. Sci. 2021, 49, 78–91. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.; Fang, H.; Zhang, T.; Su, M.; Xu, L.; Zhang, Z.; Chen, X. Nitrogen affects anthocyanin biosynthesis by regulating MdLOB52 downstream of MdARF19 in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). J. Plant Growth Regul. 2018, 37, 719–729. [Google Scholar] [CrossRef]
- Oraei, M.; Panahirad, S.; Zaare-Nahandi, F.; Gohari, G. Pre-véraison treatment of salicylic acid to enhance anthocyanin content of grape (Vitis vinifera L.) berries. J. Sci. Food Agric. 2019, 99, 5946–5952. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, L.; Yuan, C.; Guan, J. Molecular characterization of ethylene-regulated anthocyanin biosynthesis in plums during fruit ripening. Plant Mol. Biol. Report. 2016, 34, 777–785. [Google Scholar] [CrossRef]
- Tijero, V.; Teribia, N.; Munné-Bosch, S. Hormonal cross-talk in the regulation of ripening and over-ripening in sweet cherries. In Proceedings of the Conference: IX Simpósio Ibérico de Maturação e Pós-Colheita, Lisbon, Portugal, 2–4 November 2016; Volume: Actas Portuguesas de Horticultura nº 28. [Google Scholar]
- Li, J.; Liu, B.; Li, X.; Li, D.; Han, J.; Zhang, Y.; Ma, C.; Xu, W.; Wang, L.; Jiu, S. Exogenous abscisic acid mediates berry quality improvement by altered endogenous plant hormones level in “Ruiduhongyu” grapevine. Front. Plant Sci. 2021, 12, 739964. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Liu, W.; Ni, S.; Li, J. Transcriptome Analysis Reveals the Molecular Mechanisms Associated with Flower Color Formation in Camellia japonica ‘Joy Kendrick’. Forests 2023, 14, 69. https://doi.org/10.3390/f14010069
Yu S, Liu W, Ni S, Li J. Transcriptome Analysis Reveals the Molecular Mechanisms Associated with Flower Color Formation in Camellia japonica ‘Joy Kendrick’. Forests. 2023; 14(1):69. https://doi.org/10.3390/f14010069
Chicago/Turabian StyleYu, Suhang, Weixin Liu, Sui Ni, and Jiyuan Li. 2023. "Transcriptome Analysis Reveals the Molecular Mechanisms Associated with Flower Color Formation in Camellia japonica ‘Joy Kendrick’" Forests 14, no. 1: 69. https://doi.org/10.3390/f14010069




