Comparative Physiological, Transcriptomic, and Metabolomic Analyses of Acacia mangium Provide New Insights into Its Molecular Mechanism of Self-Incompatibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Samples
2.2. Microscopy Observation for Evaluate the SI Type
2.3. Total RNA Extraction and RNA-seq Analysis
2.4. UPLC-MS/MS-Based Metabolomic Analysis
2.5. Joint Transcriptomic–Metabolomic Joint Analysis of the Molecular Mechanism of SI
3. Results
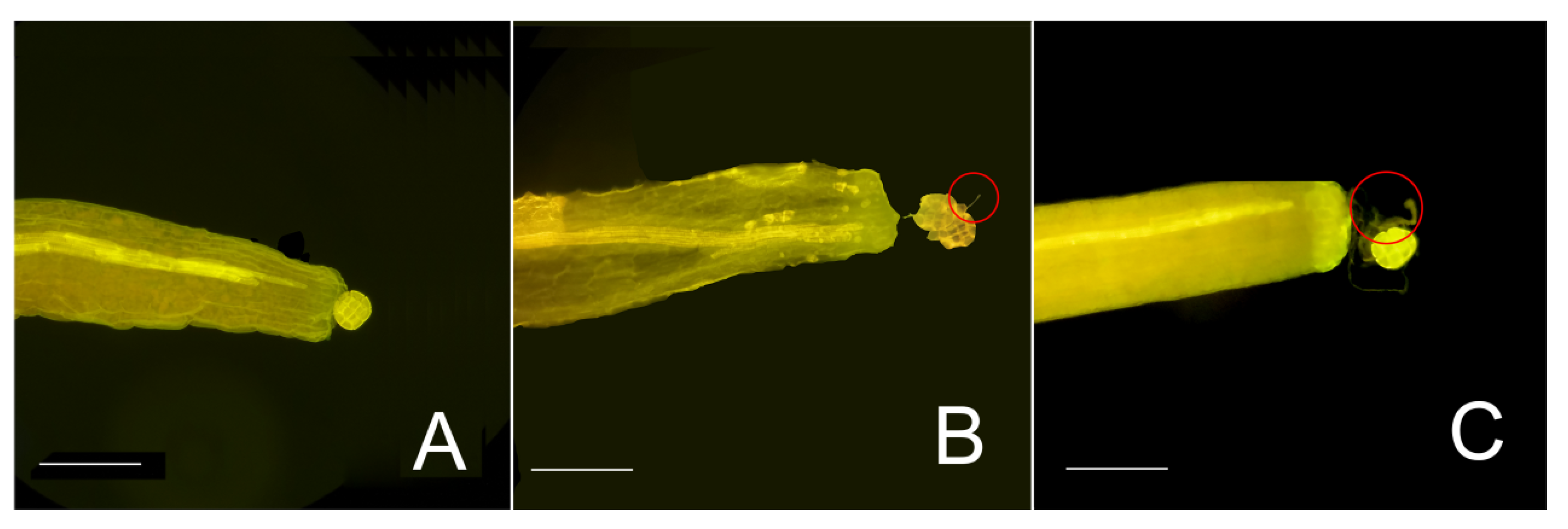
3.1. Microscopy Observation of Pollen Germination and Stigma Contact Sites
3.2. Transcriptomic Analysis
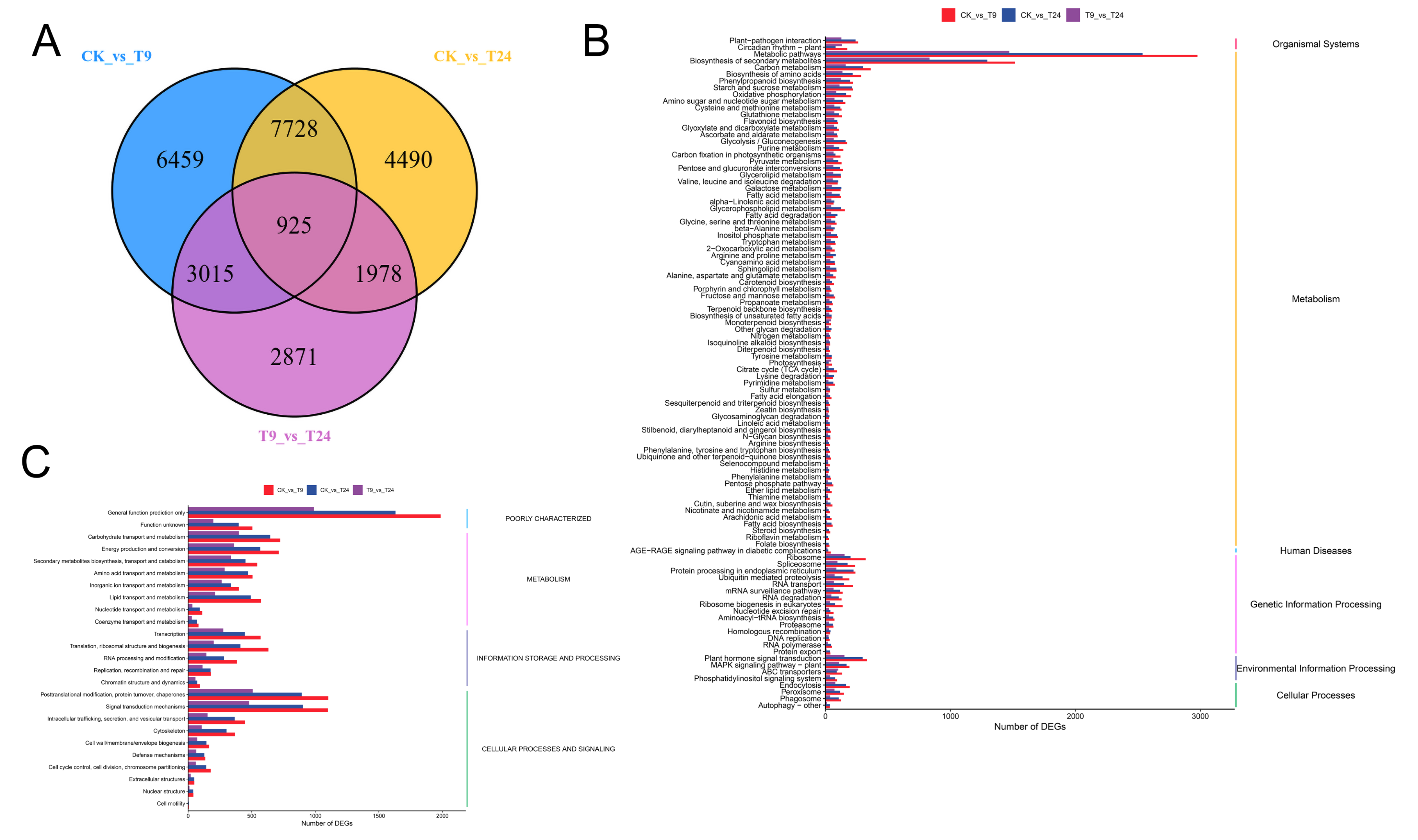
3.2.1. Transcriptome Data and Differentially Expressed Gene (DEG) Screening
3.2.2. DEGs Related to SI Genes and Growth of Pollen Tubes
3.2.3. The Co-Expression Network between Differentially Expressed TFs and SI-Related Genes
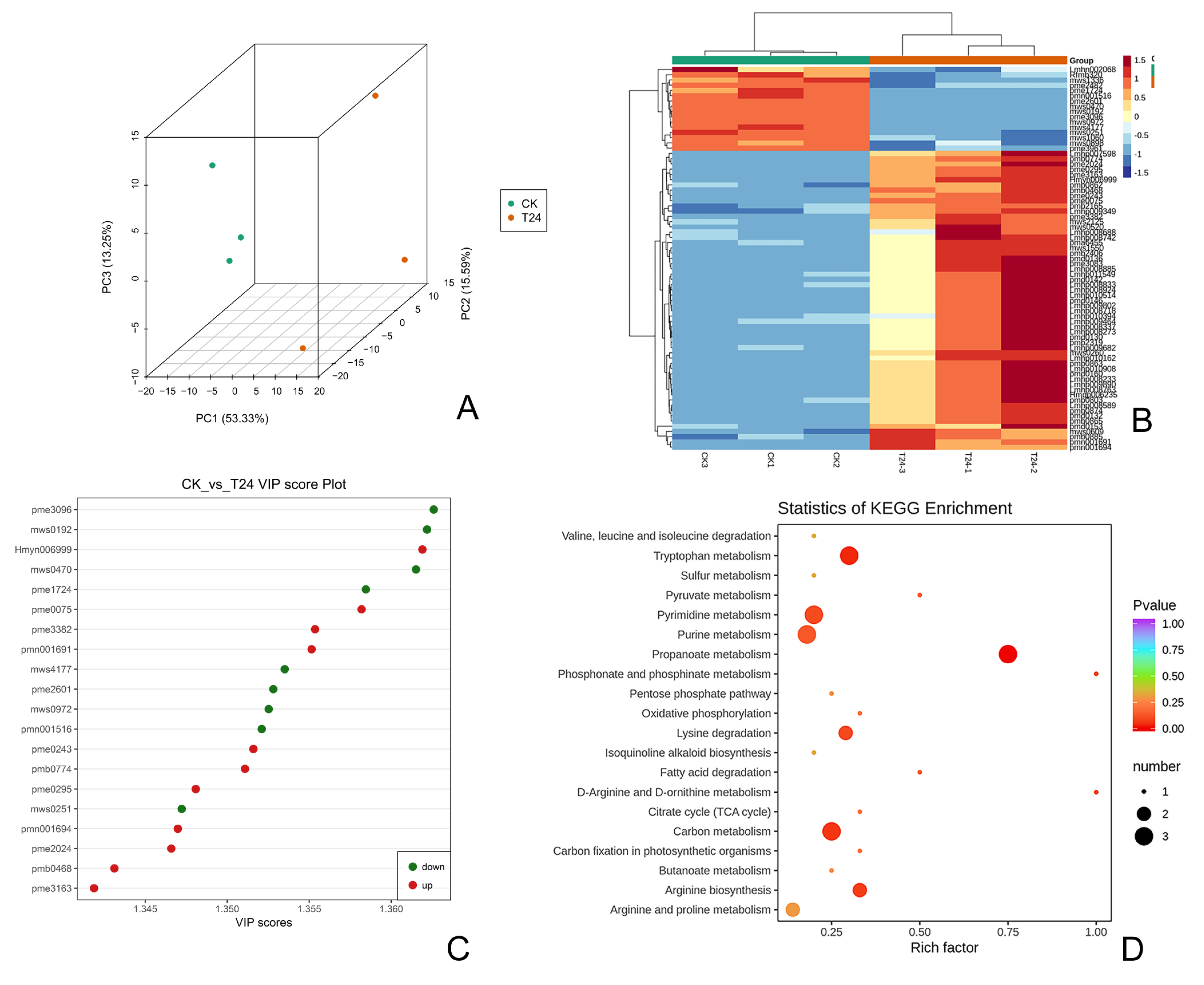
3.3. Transcriptome–Metabolome Joint Analysis
3.4. Joint Transcriptomic–Metabolomic Analysis
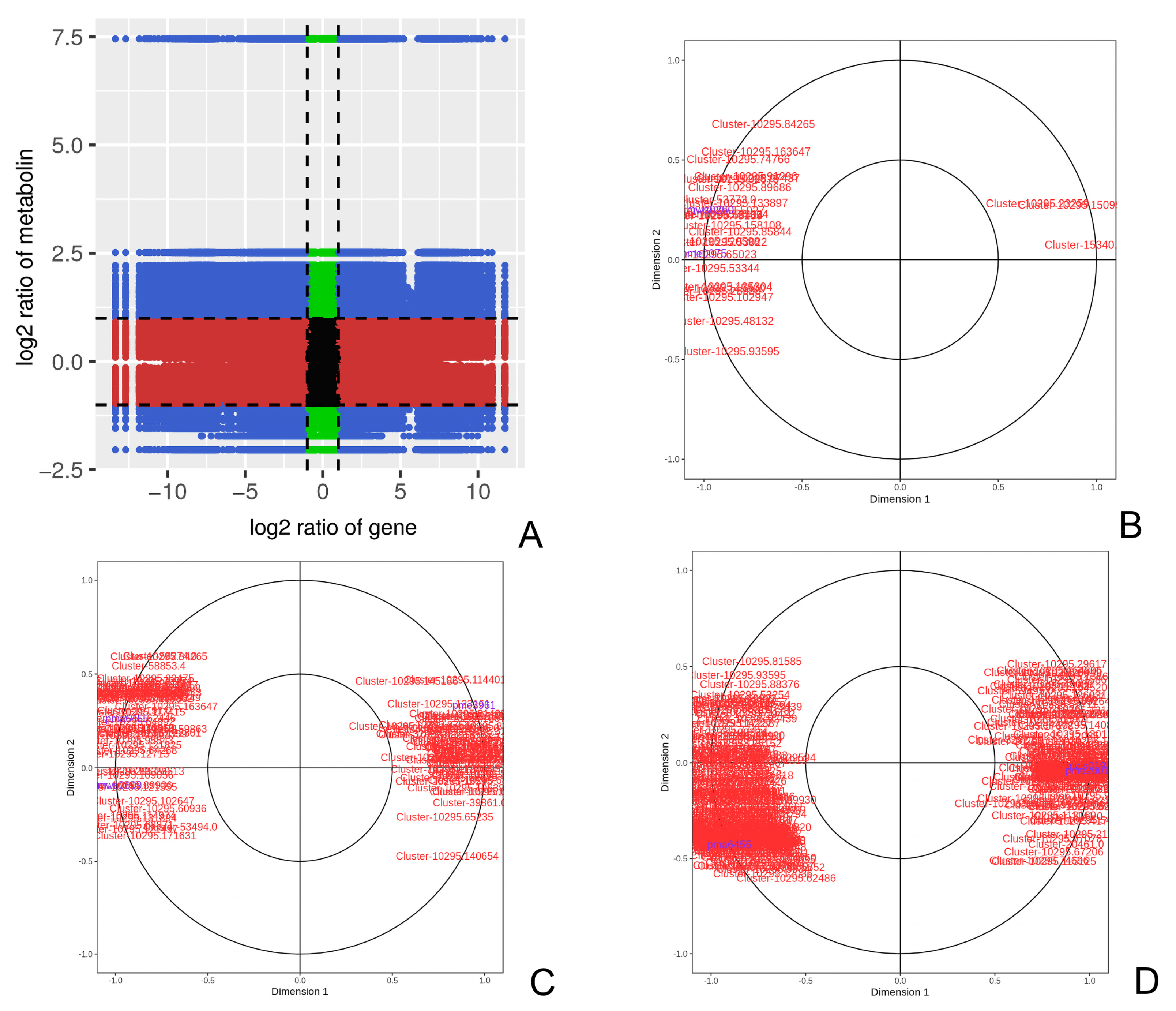
3.4.1. Differentially Expressed Genes and Metabolites Related to SI
3.4.2. Joint Analysis of the Path Graph of SI from the Perspective of Transcriptomics and Metabolomics
4. Discussion
4.1. SI Type in A. Mangium
4.2. Transcriptomic Analysis
4.3. Metabolomic Analysis
4.4. Model-Based Prediction of Transcripts and Metabolites Related to SI of A. Mangium
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Allen, A.M.; Hiscock, S.J. Evolution and Phylogeny of Self-Incompatibility Systems in Angiosperms. In Self-Incompatibility in Flowering Plants; Springer: Berlin/Heidelberg, Germany, 2008; pp. 73–101. [Google Scholar] [CrossRef]
- Liu, L. Rapid identification of self-incompatibility in radish(Raphanus sativus L.) with fluorescent observation. J. Nanjing Agric. Univ. 2007, 30, 30–34. [Google Scholar] [CrossRef]
- Jun, S.G.; Lin, H.X. Measurement of Self-incompatible by Fluoroscope Observation in Non-heading Chinese Cabbage. J. Wuhan Bot. Res. 2004, 22, 197–200. [Google Scholar]
- Jordan, N.D.; Ride, J.P.; Rudd, J.J.; Davies, E.M.; Franklin-Tong, V.E.; Franklin, F. Inhibition of Self-incompatible Pollen in Papaver rhoeas Involves a Complex Series of Cellular Events. Ann. Bot. 2000, 85, 197–202. [Google Scholar] [CrossRef][Green Version]
- Wachira, F.N.; Kamunya, S.K. Pseudo-self-incompatibility in some tea clones (Camellia sinensis (L.) O. Kuntze). J. Hortic. Sci. Biotechnol. 2005, 80, 716–720. [Google Scholar] [CrossRef]
- Chen, X.; Hao, S.; Wang, L.; Fang, W.; Wang, Y.; Li, X. Late-acting self-incompatibility in tea plant (Camellia sinensis). Biologia 2012, 67, 347–351. [Google Scholar] [CrossRef]
- Zhang, C.C.; Wang, L.Y.; Wei, K.; Wu, L.Y.; Li, H.L.; Zhang, F.; Cheng, H.; Ni, D.J. Transcriptome analysis reveals self-incompatibility in the tea plant (Camellia sinensis) might be under gametophytic control. BMC Genom. 2016, 17, 359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lu, M.; Yu, S.; Liu, Y.; Yang, J.; Tan, X. In-depth Understanding of Camellia oleifera Self-incompatibility by Comparative Transcriptome, Proteome and Metabolome. Int. J. Mol. Sci. 2020, 21, 1600. [Google Scholar] [CrossRef]
- Naithani, S.; Chookajorn, T.; Ripoll, D.R.; Nasrallah, J.B. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 2007, 104, 12211–12216. [Google Scholar] [CrossRef]
- Stein, J.C.; Howlett, B.; Boyes, D.C.; Nasrallah, N.J.B. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 1991, 88, 8816–8820. [Google Scholar] [CrossRef]
- Haffani, Y.Z.; Silva, N.F.; Goring, D.R. Receptor kinase signalling in plants. Can. J. Bot. 2004, 82, 1–15. [Google Scholar] [CrossRef][Green Version]
- Ivanov, R.; Gaude, T. Endocytosis and Endosomal Regulation of the S-Receptor Kinase during the Self-Incompatibility Response in Brassica oleracea. Plant Cell Online 2009, 21, 2107–2117. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Tong, V.E.; Holdaway-Clarke, T.L.; Straatman, K.R.; Kunkel, J.G.; Hepler, P.K. Involvement of extracellular calcium influx in the self-incompatibility response of Papaver rhoeas. Plant J. 2002, 29, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Tong, N.; Franklin, F. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 2003, 8, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Tong, V.E.; Franklin, F.C. The different mechanisms of gametophytic self-incompatibility. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2003, 358, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Blanchoin, L.; Chaudhry, F.; Franklin-Tong, V.E.; Staiger, C.J. A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 2004, 279, 23364–23375. [Google Scholar] [CrossRef]
- Liu, S.L.; Zhao, G.J.; Xin, W.U.; Zhang, B.H.; Gao, L.W.; Ding, M.L.; Chen, W. Research Progress on Plant Self-incompatibility Mechanism. J. Agric. Sci. Technol. 2016, 18, 31–37. [Google Scholar] [CrossRef]
- McClure, B. New views of S-RNase-based self-incompatibility. Curr. Opin. Plant Biol. 2006, 9, 639–646. [Google Scholar] [CrossRef]
- Sun, L.; Williams, J.S.; Li, S.; Wu, L.; Khatri, W.A.; Stone, P.G.; Keebaugh, M.D.; Kao, T.H. S-Locus F-Box Proteins Are Solely Responsible for S-RNase-Based Self-Incompatibility of Petunia Pollen. Plant Cell 2018, 30, 2959–2972. [Google Scholar] [CrossRef]
- Daele, I.V.; Bockstaele, E.V.; Martens, C.; Roldán-Ruiz, I. Identification of transcribed derived fragments involved in self-incompatibility in perennial ryegrass (Lolium perenne L.) using cDNA-AFLP. Euphytica 2008, 163, 67–80. [Google Scholar] [CrossRef]
- Rohini, G.; Patel, R.K.; Tyagi, A.K.; Mukesh, J. De Novo Assembly of Chickpea Transcriptome Using Short Reads for Gene Discovery and Marker Identification. DNA Res. 2011, 18, 53–63. [Google Scholar] [CrossRef]
- He, Y.; Song, Q.; Wu, Y.; Ye, S.; Chen, S.; Chen, H. TMT-Based Quantitative Proteomic Analysis Reveals the Crucial Biological Pathways Involved in Self-Incompatibility Responses in Camellia oleifera. Int. J. Mol. Sci. 2020, 21, 1987. [Google Scholar] [CrossRef]
- Seif, A.; Wang, P.; Li, N.; Chen, Y.; Lin, S. Metabolomic Analysis of Pollen Grains with Different Germination Abilities from Two Clones of Chinese Fir (Cunninghamia lanceolata (Lamb) Hook). Molecules 2018, 23, 3162. [Google Scholar] [CrossRef]
- Kenrick, J.; Knox, R.B. Quantitative Analysis of Self-Incompatibility in Trees of Seven Species of Acacia. J. Hered. 1989, 80, 240–245. [Google Scholar] [CrossRef]
- Kho, Y.O.; Bar, J. Observing pollen tubes by means of fluorescence. Euphytica 1968, 17, 298–302. [Google Scholar] [CrossRef]
- Gao, C.; Yuan, D.; Yang, Y.; Wang, B.; Liu, D.; Feng, Z.; Tan, X. Anatomical characteristics of self-incompatibility of Camellia oleifera. Sci. Silvae Sin. 2015, 51, 60–68. [Google Scholar]
- Kenrick, J.; Williams, V.K.G. Self-incompatibility in Acacia retinodes: Site of pollen-tube arrest is the nucellus. Planta 1986, 169, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Shivanna, K.R.; Ram, H.M. Pollination biology and breeding system of Acacia senegal. Biol. J. Linn. Soc. 2000, 135, 251–262. [Google Scholar] [CrossRef]
- Li, Z.; Lin, Y.; Peng, S. Nutrient content in litterfall and its translocation in plantation forests in south China. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2000, 11, 321–326. [Google Scholar] [CrossRef]
- Kumar, A.; Jhariya, M.K.; Yadav, D.K.; Banerjee, A. Vegetation dynamics in Bishrampur collieries of northern Chhattisgarh, India: Eco-restoration and management perspectives. Environ. Monit. Assess. 2017, 189, 371. [Google Scholar] [CrossRef] [PubMed]
- Caldeira, M.V.W.; Favalessa, M.; Delarmelina, W.M.; Gonçalves, E.D.O.; Santos Moura, R.R.D. Sewage sludge assessment on growth of Acacia mangium seedlings by principal components analysis and orthogonal contrasts. J. Plant Nutr. 2018, 41, 1303–1311. [Google Scholar] [CrossRef]
- Chen, J.; Shen, W.; Xu, H.; Li, Y.; Luo, T. The Composition of Nitrogen-Fixing Microorganisms Correlates with Soil Nitrogen Content during Reforestation: A Comparison between Legume and Non-legume Plantations. Front. Microbiol. 2019, 10, 508. [Google Scholar] [CrossRef]
- Silva, J.L.; Demolin Leite, G.L.; de Souza Tavares, W.; Souza Silva, F.W.; Sampaio, R.A.; Azevedo, A.M.; Serrão, J.E.; Zanuncio, J.C. Diversity of arthropods on Acacia mangium (Fabaceae) and production of this plant with dehydrated sewage sludge in degraded area. R. Soc. Open Sci. 2020, 7, 191196. [Google Scholar] [CrossRef]
- Lima, J.S.; Leite, G.L.D.; Guanabens, P.F.S.; Soares, M.A.; Silva, J.L.; Mota, M.V.S.; Lemes, P.G.; Zanuncio, J.C. Insects and spiders on Acacia mangium (Fabaceae) saplings as bioindicators for the recovery of tropical degraded areas. Braz. J. Biol. Rev. Bras. Biol. 2021, 84, e252088. [Google Scholar] [CrossRef]
- Zeng, S.C.; Su, Z.Y.; Gu, Y.K.; Xie, Z.S.; Liu, Y.X. Litterfalls of major forest stands at Baiyunshan scenic spot of Guangzhou. Chin. J. Appl. Ecol. 2003, 14, 3. [Google Scholar] [CrossRef]
- Pinto, P.C.; Evtuguin, D.V.; Neto, C.P. Chemical composition and structural features of the macromolecular components of plantation Acacia mangium wood. J. Agric. Food Chem. 2005, 53, 7856–7862. [Google Scholar] [CrossRef] [PubMed]
- Hermawan, D.; Hadi, Y.S.; Fajriani, E.; Massijaya, M.Y.; Hadjib, N. Resistance of Particleboards Made from Fast-Growing Wood Species to Subterranean Termite Attack. Insects 2012, 3, 532–537. [Google Scholar] [CrossRef]
- Wang, Z.J.; Ye, G.F.; Tan, F.L.; Chen, S. The study on stress resistance of acacia tree species in coastal sandy site. J. Fujian For. Sci. Technol. 2005, 4, 35–38. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; Zhao, P.; Ni, G.Y.; Zhu, L.W.; Zhao, X.H.; Zhao, P.Q.; Niu, J.F. Water use of re-vegetation pioneer tree species Schima superba and Acacia mangium in hilly land of South China. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2014, 25, 931–939. [Google Scholar]
- Hegde, N.; Joshi, S.; Soni, N.; Kushalappa, A.C. The caffeoyl-CoA O-methyltransferase gene SNP replacement in Russet Burbank potato variety enhances late blight resistance through cell wall reinforcement. Plant Cell Rep. 2021, 40, 237–254. [Google Scholar] [CrossRef]
- Huang, L.J.; Zhan, N.; Li, J. Floral biology of Acacia mangium. For. Res. 2014, 27, 45–52. [Google Scholar] [CrossRef]
- Zhan, N.; Huang, L.J.; Jun, L.I. Study the Reasons of Low Pod of Acacia mangium Seed Orchard. J. Southwest Univ. (Nat. Sci. Ed.) 2016, 38, 46–52. [Google Scholar] [CrossRef]
- Zhang, R.P.; Zeng, B.S.; Huang, L.J. Preliminary study on hydroponics culture of flower branch of A. mangium and A. auriculiformis. Acta Agric. Shanghai 2021, 37, 70–73. [Google Scholar] [CrossRef]
- Zhan, N.; Huang, L.J. Study on Acacia mangium pollen germination in vitro. Guihaia 2016, 36, 92–96+125. [Google Scholar] [CrossRef]
- Zhang, R.P.; Zhan, N.; Huang, L.J. Control Pollination of Acacia mangium and A. auriculiformis. Mol. Plant Breed. 2020, 18, 5877–5882. [Google Scholar] [CrossRef]
- Zhan, N. Three Acacia Pollen Vigor Test and Controlled Pollination. Mater’s Thesis, Chinese Academy of Forestry, Beijing, China, 2016. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Gueguen, L.; Coppee, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Alcaraz, N.; Friedrich, T.; Kötzing, T.; Krohmer, A.; Müller, J.; Pauling, J.; Baumbach, J. Efficient key pathway mining: Combining networks and OMICS data. Integr. Biol. Quant. Biosci. Nano Macro 2012, 4, 756–764. [Google Scholar] [CrossRef]
- Alcaraz, N.; Pauling, J.; Batra, R.; Barbosa, E.; Junge, A.; Christensen, A.G.; Azevedo, V.; Ditzel, H.J.; Baumbach, J. KeyPathwayMiner 4.0: Condition-specific pathway analysis by combining multiple omics studies and networks with Cytoscape. BMC Syst. Biol. 2014, 8, 99. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bylesjö, M.; Eriksson, D.; Kusano, M.; Moritz, T.; Trygg, J. Data integration in plant biology: The O2PLS method for combined modeling of transcript and metabolite data. Plant J. 2007, 52, 1181–1191. [Google Scholar] [CrossRef]
- Hong, S.; Chen, X.; Jin, L.; Xiong, M. Canonical correlation analysis for RNA-seq co-expression networks. Nucleic Acids Res. 2013, 41, e95. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.H.; Xiaomin, L. Self-incompatibility in Poaceae and its Advancement in Molecular Biological Research. Mol. Plant Breed. 2006, 4, 269–274. [Google Scholar] [CrossRef]
- Nettancourt, D. Incompatibility and Incongruity in Wild and Cultivated Plants; Springer: Berlin/Heidelberg, Germany, 2001; pp. 15–24. [Google Scholar] [CrossRef]
- Li, F.; Tang, F.; Gao, D.; Duan, S.; Li, Y.; Li, C.; Ma, L. Research progress on gametophytic self-incompatibility. J. Yunnan Norm. Univ. (Nat. Sci. Ed.) 2019, 39, 65–70. [Google Scholar]
- Luis, B.J.; Cocucci, A.A.; Anton, A.M. Reproductive biology in Acacia caven (Mol.) Mol. (Leguminosae) in the central region of Argentina. Bot. J. Linn. Soc. 1995, 119, 65–76. [Google Scholar] [CrossRef]
- Sedgley, M.; Harbard, J.; Smith, R.M.M.; Wickneswari, R.; Griffin, A.R. Reproductive-Biology and Interspecific Hybridization of Acacia mangium and Acacia auriculiformis A. Cunn. ex Benth (Leguminosae, Mimosoideae). Aust. J. Bot. 1992, 40, 37–48. [Google Scholar] [CrossRef]
- Wehling, P.; Hackauf, B.; Wricke, G. Phosphorylation of pollen proteins in relation to self-incompatibility in rye (Secale cereale L.). Sex. Plant Reprod. 1994, 7, 67–75. [Google Scholar] [CrossRef]
- Rudd, J.J. Ca2+—Independent phosphorylation of a 68 kDa pollen protein is stimulated by the self-incompatibility response in Papaver rhoeas. Plant J. 2010, 12, 507–514. [Google Scholar] [CrossRef]
- Peralta, I.; Rodríguez, J.; Arroyo, M. Breeding system and aspects of pollination in Acacia caven (Mol.) Mol. (Leguminosae: Mimosoideae) in the mediterranean-type climate zone of central Chile. Bot. Jarbücher Für Syst. Pflanzengesch. Pflanzengeogr. 1992, 114, 297–314. [Google Scholar]
- Klaas, M.; Yang, B.; Bosch, M.; Thorogood, D.; Manzanares, C.; Armstead, I.P.; Franklin, F.C.H. Progress towards elucidating the mechanisms of self-incompatibility in the grasses: Further insights from studies in Lolium. Ann. Bot. 2011, 108, 677. [Google Scholar] [CrossRef]
- Zhan, N.; Huang, L.J. Effects of Ca2+ on in vitro pollen germination of three Acacia species. Silvae Genet. 2016, 65, 11–16. [Google Scholar] [CrossRef][Green Version]
- Rogel, M.R.; Jaitovich, A.; Ridge, K.M. The role of the ubiquitin proteasome pathway in keratin intermediate filament protein degradation. Proc. Am. Thorac. Soc. 2010, 7, 71–76. [Google Scholar] [CrossRef]
- Shang, F.; Taylor, A. Roles for the ubiquitin-proteasome pathway in protein quality control and signaling in the retina: Implications in the pathogenesis of age-related macular degeneration. Mol. Asp. Med. 2012, 33, 446–466. [Google Scholar] [CrossRef]
- Kao, T.H.; Tsukamoto, T. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 2004, 16, S72–S83. [Google Scholar] [CrossRef]
- Kakeda, K. S locus-linked F-box genes expressed in anthers of Hordeum bulbosum. Plant Cell Rep. 2009, 28, 1453–1460. [Google Scholar] [CrossRef]
- Wei, L.; Li, P.K.; Zhang, H.Q.; Wang, T.C. Research progress in plant thioredoxin. Guizhou Agric. Sci. 2006, 129–131, 123. [Google Scholar] [CrossRef]
- Li, X.; Paech, N.A.; Nield, J.; Hayman, D.; Langridge, P. Self-incompatibility in the grasses: Evolutionary relationship of the S gene from Phalaris coerulescens to homologous sequences in other grasses. Plant Mol. Biol. 1997, 34, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Díaz, J.A.; McClure, B.; Vázquez-Santana, S.; Guevara-García, A.; León-Mejía, P.; Márquez-Guzmán, J.; Cruz-García, F. A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNase in vitro. J. Biol. Chem. 2006, 281, 3418–3424. [Google Scholar] [CrossRef]
- Torres-Rodríguez, M.D.; González-Segura, L.; Rodríguez-Sotres, R.; Juárez-Día, Z.J.; Cruz-Zamora, Y.; Cruz-García, F. High resolution crystal structure of NaTrxh from Nicotiana alata and its interaction with the S-RNase. J. Struct. Biol. 2020, 212, 107578. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Song, Y.; Zhang, H.; Fan, J.; Li, Q.; Zhang, D.; Xue, Y. Electrostatic potentials of the S-locus F-box proteins contribute to the pollen S specificity in self-incompatibility in Petunia hybrida. Plant J. 2017, 89, 45–57. [Google Scholar] [CrossRef]
- Li, Y.; Duan, X.; Wu, C.; Yu, J.; Liu, C.; Wang, J.; Zhang, X.; Yan, G.; Jiang, F.; Li, T.; et al. Ubiquitination of S(4)-RNase by S-LOCUS F-BOX LIKE2 Contributes to Self-Compatibility of Sweet Cherry ‘Lapins’. Plant Physiol. 2020, 184, 1702–1716. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Q.; Zhang, X.; Wang, J.; Ayup, M.; Yang, B.; Guo, C.; Gong, P.; Dong, W. The proteome reveals the involvement of serine/threonine kinase in the recognition of self-incompatibility in almond. J. Proteom. 2022, 256, 104505. [Google Scholar] [CrossRef]
- Burrows, B.A.; McCubbin, A.G. Sequencing the genomic regions flanking S-linked PvGLO sequences confirms the presence of two GLO loci, one of which lies adjacent to the style-length determinant gene CYP734A50. Plant Reprod. 2017, 30, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Qin, Y.; Silva, J.A.T.D.; Ye, Z.; Hu, G. Identification of differentially expressed genes in pistils from self-incompatible Citrus reticulata by suppression subtractive hybridization. Mol. Biol. Rep. 2013, 40, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Jia, J.; Huang, X.; Yan, X.; Cheng, L.; Chen, S.; Li, X.; Peng, X.; Liu, G. The large-scale investigation of gene expression in Leymus chinensis stigmas provides a valuable resource for understanding the mechanisms of poaceae self-incompatibility. BMC Genom. 2014, 15, 399. [Google Scholar] [CrossRef][Green Version]
- Heizmann, P.; Luu, D.T.; Dumas, C. Pollen-Stigma Adhesion in the Brassicaceae. Ann. Bot. 2000, 85, 23–27. [Google Scholar] [CrossRef]
- Ischebeck, T. Lipids in pollen—They are different. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Nagel, C.; Taylor, L.P. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 1992, 89, 7213–7217. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Guivarćh, A.; Hirsche, J.; Bauerfeind, M.A.; González, M.C.; Hyun, T.K.; Eom, S.H.; Chriqui, D.; Engelke, T.; Großkinsky, D.K.; et al. Metabolic Control of Tobacco Pollination by Sugars and Invertases. Plant Physiol. 2017, 173, 984–997. [Google Scholar] [CrossRef]
- Zhao, P.; Pan, Q.; Yu, W.; Zhao, L. Dissect style response to pollination using metabolite profiling in self-compatible and self-incompatible tomato species. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1017, 153–162. [Google Scholar] [CrossRef]
- Hugot, K.; Ponchet, M.; Marais, A.; Ricci, P.; Galiana, E. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Mol. Plant Microbe Interact. 2002, 15, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Li, W.; Gao, S.; Lei, T.; Hu, J.; Shen, P.; Li, Y.; Li, J. Untargeted metabolomic profiling reveals that different responses to self and cross pollination in each flower morph of the heteromorphic plant Plumbago auriculata. Plant Physiol. Biochem. 2019, 144, 413–426. [Google Scholar] [CrossRef]
- Porto, N.P.; Bret, R.S.C.; Souza, P.V.L.; Cândido-Sobrinho, S.A.; Medeiros, D.B.; Fernie, A.R.; Daloso, D.M. Thioredoxins regulate the metabolic fluxes throughout the tricarboxylic acid cycle and associated pathways in a light-independent manner. Plant Physiol. Biochem. 2022, 193, 36–49. [Google Scholar] [CrossRef]
- Nakatsukasa, K.; Kawarasaki, T.; Moriyama, A. Heterologous expression and functional analysis of the F-box protein Ucc1 from other yeast species in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2019, 128, 704–709. [Google Scholar] [CrossRef]
- Lénárt, J.; Gere, A.; Causon, T.; Hann, S.; Dernovics, M.; Németh, O.; Hegedűs, A.; Halász, J. LC-MS based metabolic fingerprinting of apricot pistils after self-compatible and self-incompatible pollinations. Plant Mol. Biol. 2021, 105, 435–447. [Google Scholar] [CrossRef]

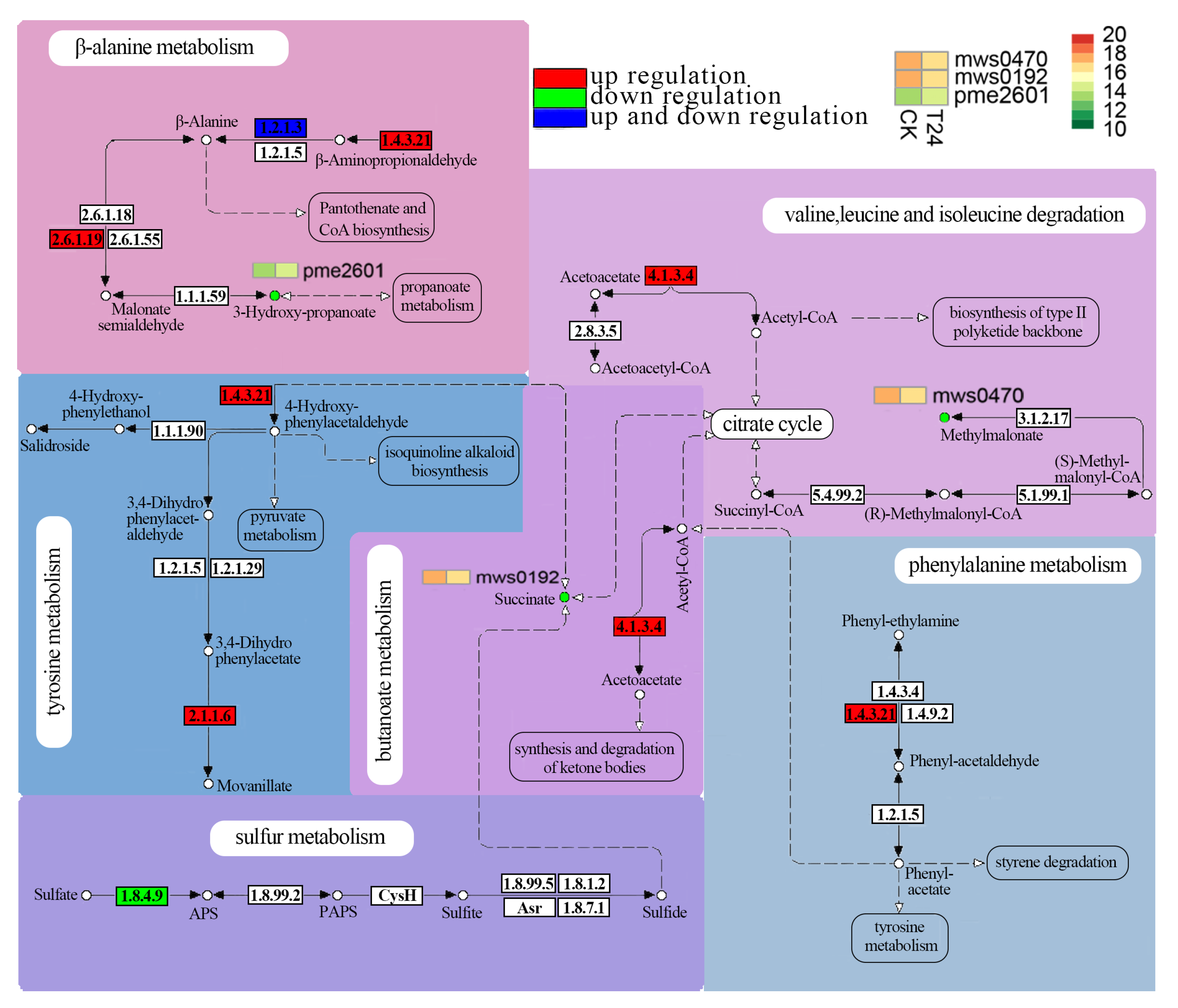
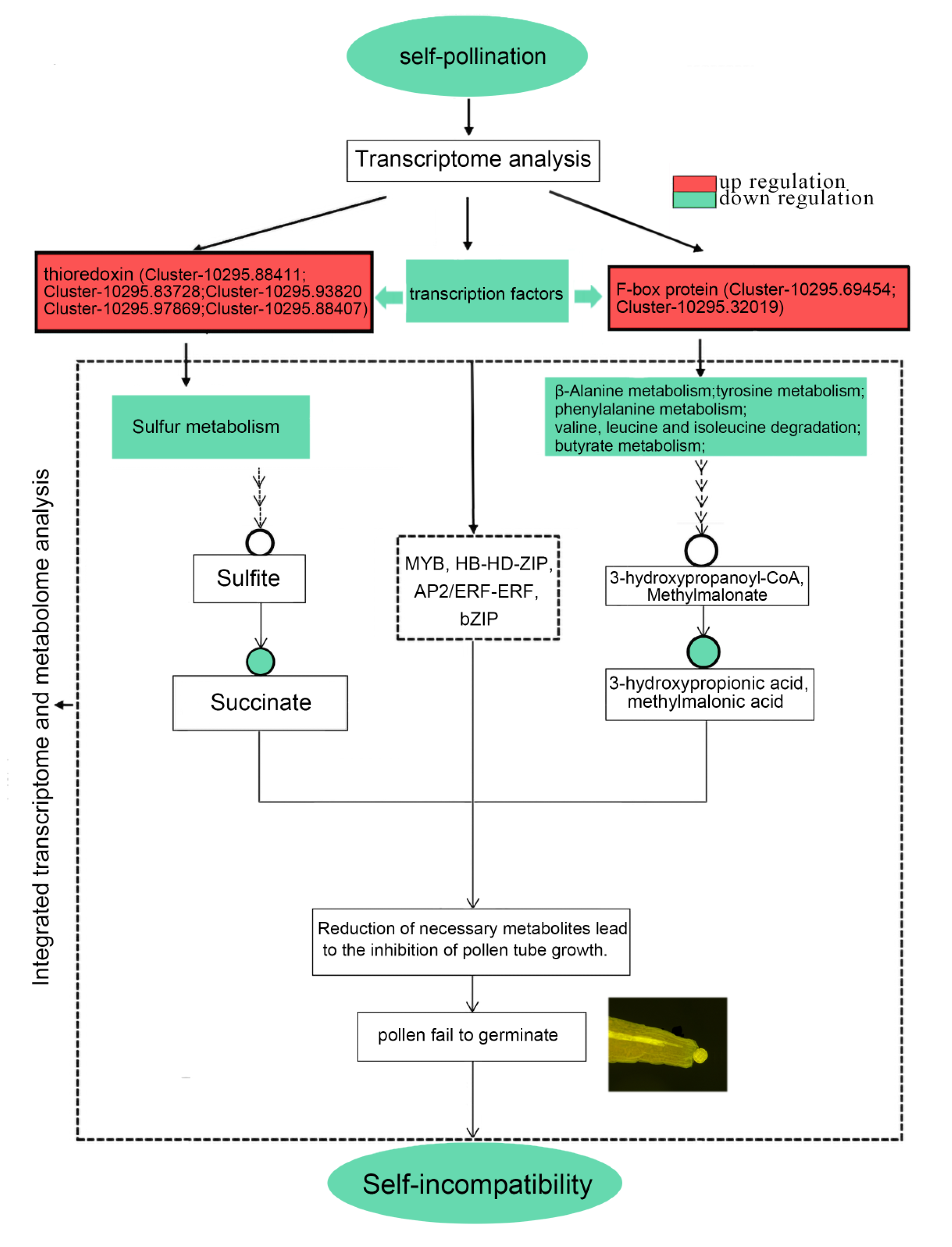
| Differential Gene Families | Differential Gene Number | Pathway ID | Pathway Annotation | Differential Metabolite |
|---|---|---|---|---|
| Thioredoxin | Cluster-10295.88411 Cluster-10295.83728 Cluster-10295.93820 Cluster-10295.97869 Cluster-10295.88407 | ko00920 | sulfur metabolism | succinate |
| F-box protein | Cluster-10295.69454 | ko00350 | tyrosine metabolism | succinate |
| ko00360 | phenylalanine metabolism | succinate | ||
| ko00410 | β-alanine metabolism | 3-hydroxypropionate | ||
| Cluster-10295.32019 | ko00280 | valine, leucine and isoleucine degradation | methylmalonate | |
| ko00650 | butanoate metabolism | succinate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Huang, L.; Zeng, B. Comparative Physiological, Transcriptomic, and Metabolomic Analyses of Acacia mangium Provide New Insights into Its Molecular Mechanism of Self-Incompatibility. Forests 2023, 14, 2034. https://doi.org/10.3390/f14102034
Zhang R, Huang L, Zeng B. Comparative Physiological, Transcriptomic, and Metabolomic Analyses of Acacia mangium Provide New Insights into Its Molecular Mechanism of Self-Incompatibility. Forests. 2023; 14(10):2034. https://doi.org/10.3390/f14102034
Chicago/Turabian StyleZhang, Ruping, Liejian Huang, and Bingshan Zeng. 2023. "Comparative Physiological, Transcriptomic, and Metabolomic Analyses of Acacia mangium Provide New Insights into Its Molecular Mechanism of Self-Incompatibility" Forests 14, no. 10: 2034. https://doi.org/10.3390/f14102034
APA StyleZhang, R., Huang, L., & Zeng, B. (2023). Comparative Physiological, Transcriptomic, and Metabolomic Analyses of Acacia mangium Provide New Insights into Its Molecular Mechanism of Self-Incompatibility. Forests, 14(10), 2034. https://doi.org/10.3390/f14102034






