Fertility Variation and Gene Diversity in Forest Populations
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Fertility Variations Estimation
3.1.1. Variations in Female and Male Fertility
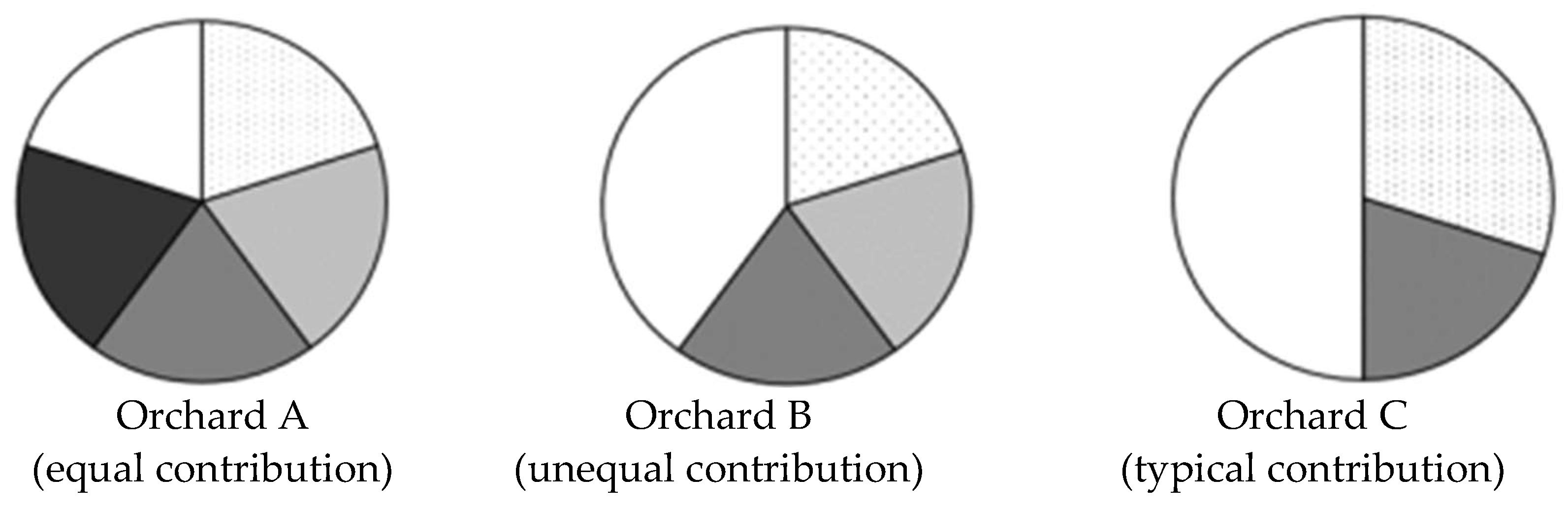
3.1.2. Total Fertility Variation (Sibling Coefficient)
3.2. Linkage of Parameters of Gene Diversity
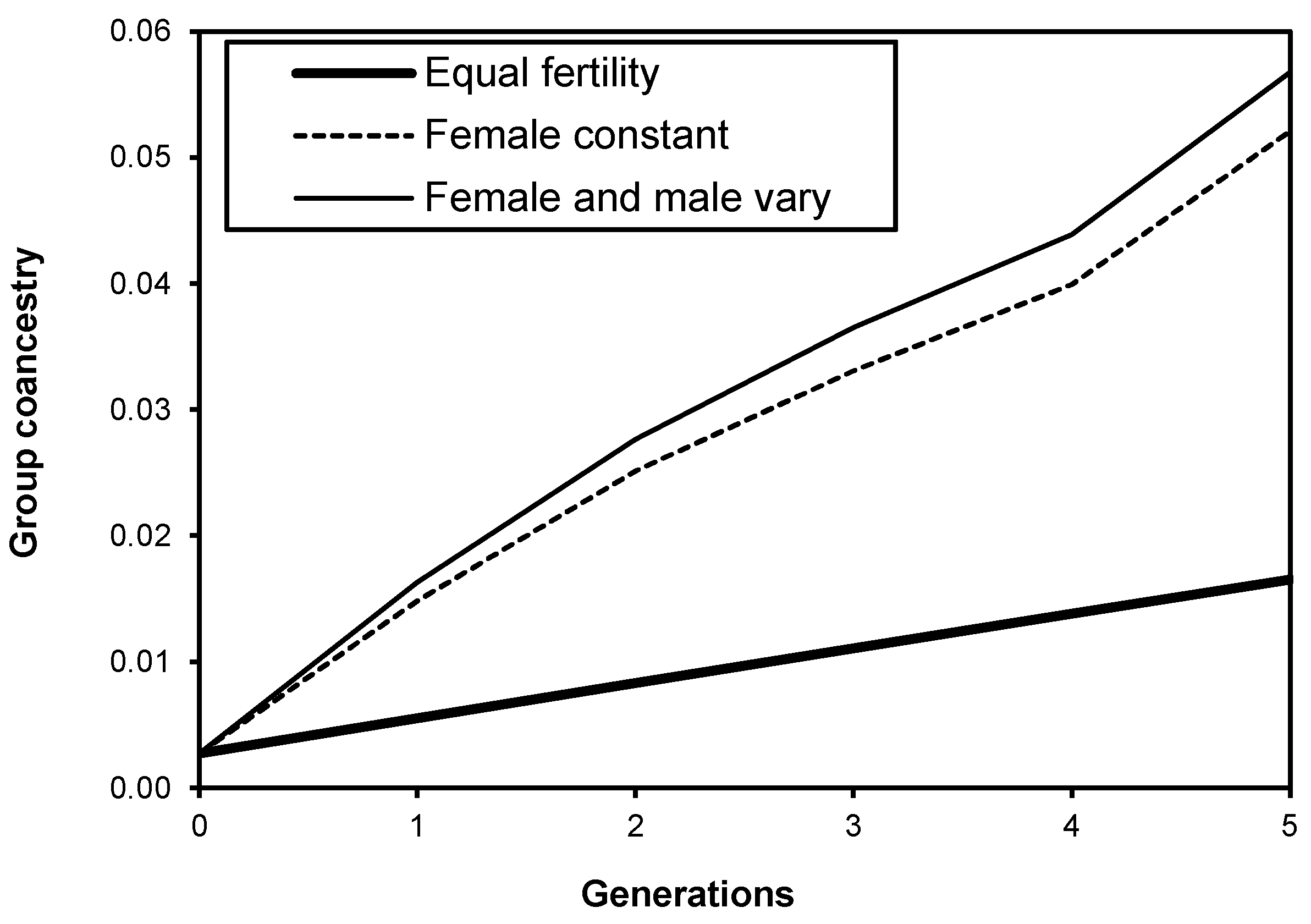
3.2.1. Coancestry and Group Coancestry
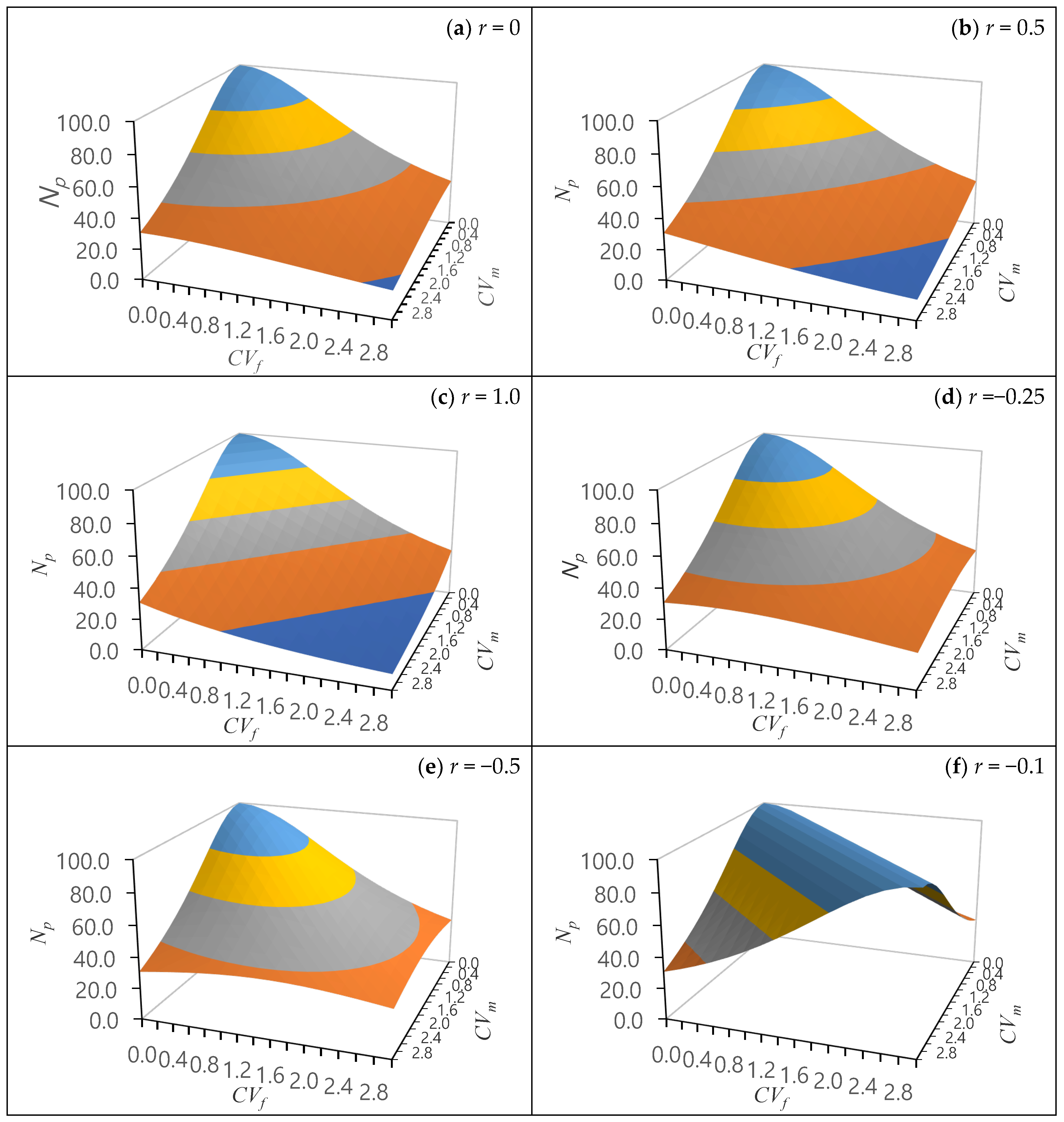
3.2.2. Gene Diversity and Status Number
3.2.3. Covariance between Female and Male Fertility
3.2.4. Sibling Coefficient and Relative Effective Number
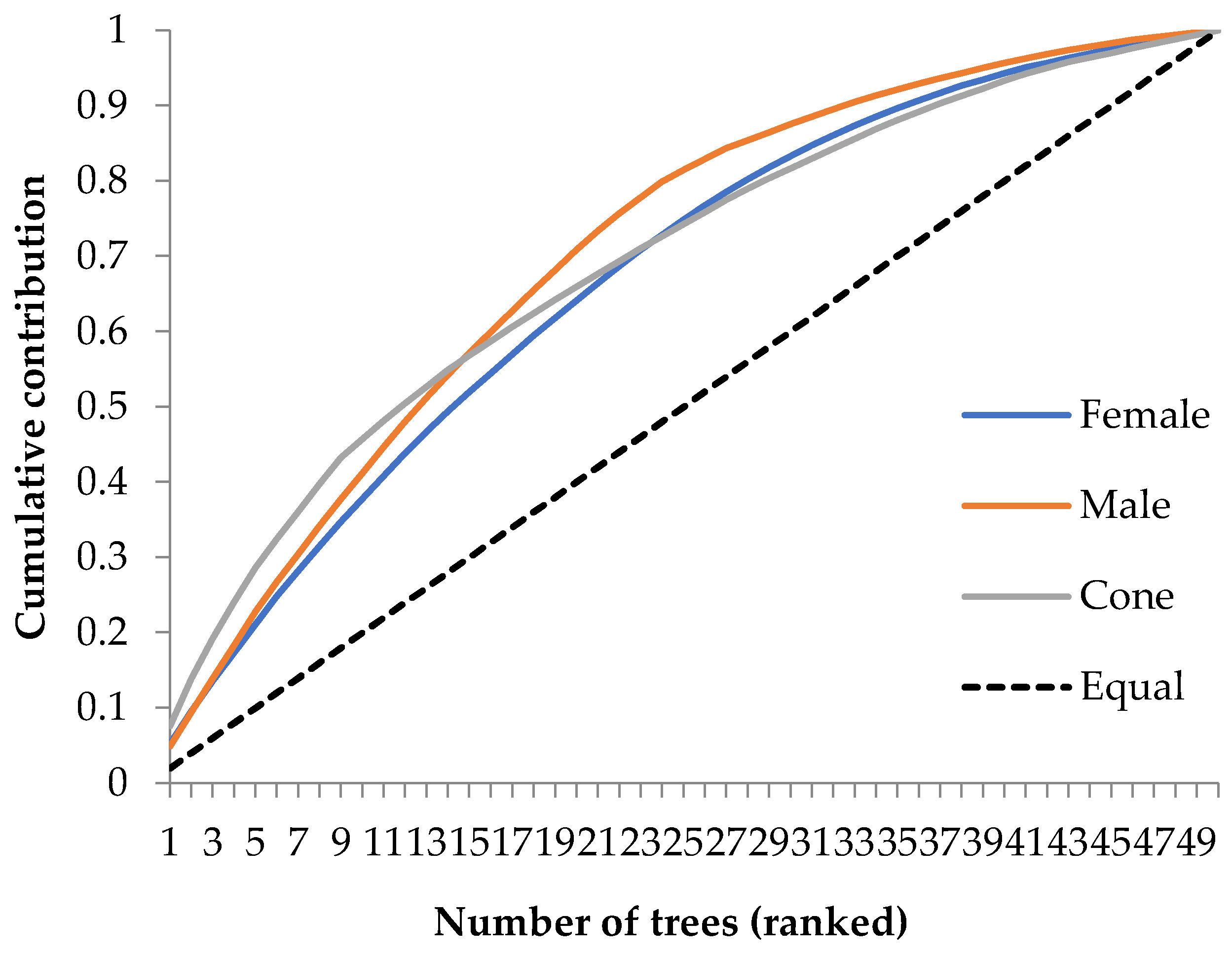
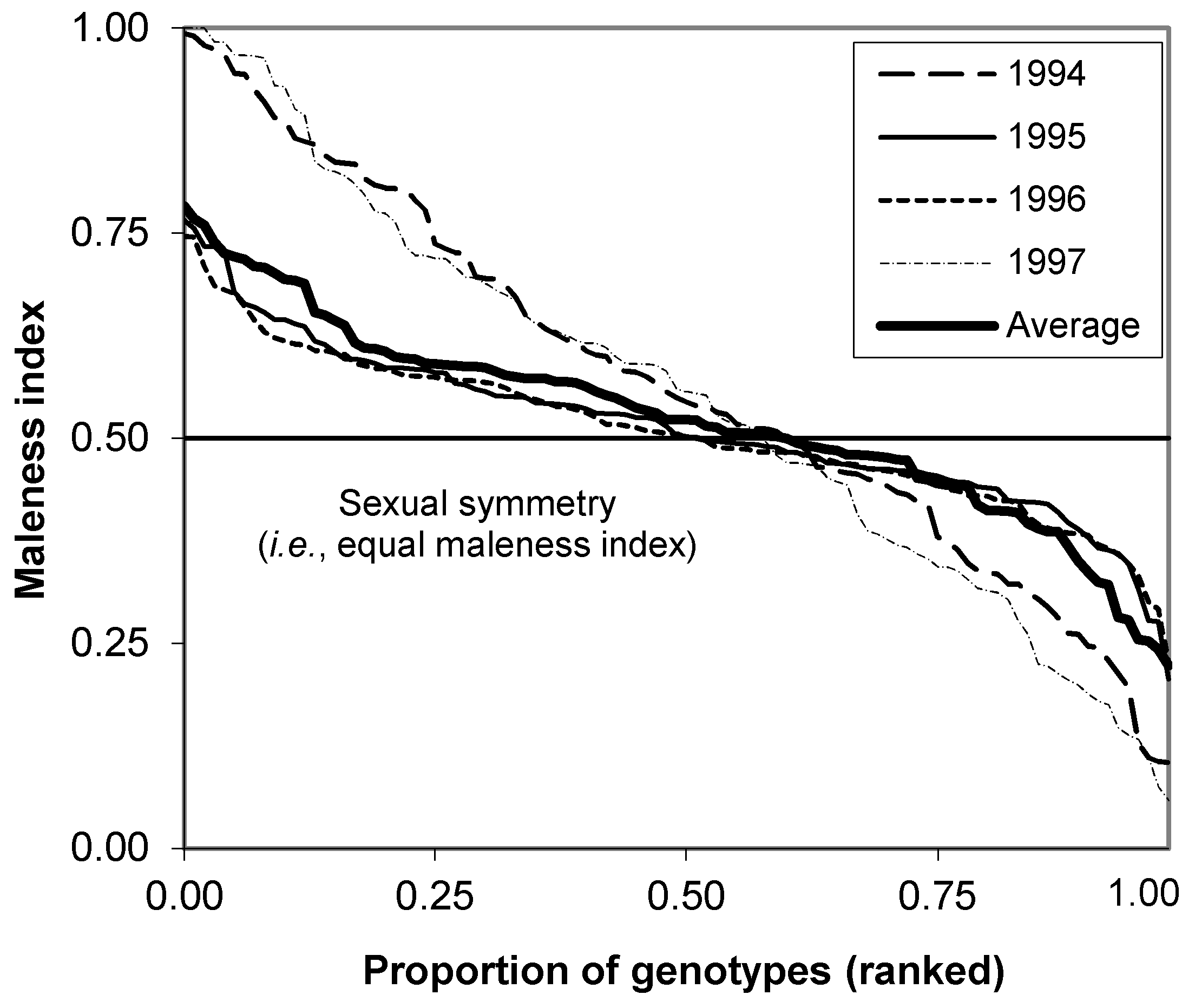
3.2.5. Parental Balance Curve and Maleness Index
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindgren, D.; Matheson, A.C. An algorithm for increasing the genetic quality of seed from seed orchards by using the better clones in higher proportions. Silvae Genet. 1986, 35, 173–177. [Google Scholar]
- Lindgren, D.; Danusevicius, D.; Rosvall, O. Unequal deployment of clones to seed orchards by considering genetic gain, relatedness and gene diversity. Forestry 2009, 82, 17–28. [Google Scholar] [CrossRef]
- Olsson, T.; Lindgren, D.; Li, B. Balancing genetic gain and relatedness in seed orchards. Silvae Genet. 2001, 50, 222–227. [Google Scholar]
- Chaisurisri, K.; El-Kassaby, Y.A. Estimation of clonal contribution to cone and seed crops in a Sitka spruce seed orchard. Ann. For. Sci. 1993, 50, 461–467. [Google Scholar] [CrossRef][Green Version]
- Kjær, E.D. Estimation of effective population number in a Picea abies seed orchard based on flower assessment. Scand. J. For. Res. 1996, 11, 111–121. [Google Scholar] [CrossRef]
- Lindgren, D.; Mullin, T.J. Relatedness and status number in seed orchard crops. Can. J. For. Res. 1998, 28, 276–283. [Google Scholar] [CrossRef]
- Reynolds, S.; El-Kassaby, Y.A. Parental balance in Douglas-fir seed orchards-cone crop vs. seed crop. Silvae Genet. 1990, 39, 40–42. [Google Scholar]
- Funda, T.; Lstibůrek, M.; Lachout, P.; Klápště, J.; El-Kassaby, Y.A. Optimization of combined genetic gain and diversity for collection and deployment of seed orchard crops. Tree Genet. Genomes 2009, 5, 583–593. [Google Scholar] [CrossRef]
- Wu, H.; Duan, A.; Wang, X.; Chen, Z.; Zhang, X.; He, G.; Zhang, J. Construction of a Core Collection of Germplasms from Chinese Fir Seed Orchards. Forests 2023, 14, 305. [Google Scholar] [CrossRef]
- Bila, A.D. Fertility Variation and Its Effects on Gene Diversity in Forest Tree Populations. Ph.D. Thesis, Swedish University of Agricultural Science, Acta Universitatis Agriculturae Sueciae, Umeå, Sweden, 2000. [Google Scholar]
- Kang, K.S. Genetic Gain and Gene Diversity of Seed Orchard Crops. Ph.D. Thesis, Swedish University of Agricultural Science, Acta Universitatis Agriculturae Sueciae, Umeå, Sweden, 2001. [Google Scholar]
- Kang, K.S.; Bila, A.D.; Harju, A.M.; Lindgren, D. Estimation of fertility variation in forest tree populations. Forestry 2003, 76, 329–344. [Google Scholar] [CrossRef]
- Bilir, N.; Kang, K.S.; Lindgren, D. Fertility variation in six populations of Brutian pine (Pinus brutia Ten.) over altitudinal ranges. Euphytica 2005, 141, 163–168. [Google Scholar] [CrossRef]
- Bilir, N.; Kang, K.S. Fertility variation, seed collection and gene diversity in natural stands of Taurus cedar (Cedrus libani). Eur. J. For. Res. 2021, 40, 199–208. [Google Scholar] [CrossRef]
- Kamalakannan, R.; Varghese, M.; Park, J.M.; Kwon, S.H.; Song, J.H.; Kang, K.S. Fertility variation and its impact on effective population size in seed stands of Tamarindus indica and Azadirachta indica. Silvae Genet. 2015, 64, 91–99. [Google Scholar] [CrossRef]
- Yazici, N.; Bilir, N. Aspectual fertility variation and its effect on gene diversity of seeds in natural stands of Taurus cedar (Cedrus libani A. Rich.). Int. J. Genom. 2017, 2017, 2960624. [Google Scholar] [CrossRef]
- Park, J.M.; Kwon, S.H.; Lee, H.J.; Na, S.J.; El-Kassaby, Y.A.; Kang, K.S. Integrating fecundity variation and genetic relatedness in estimating the gene diversity of seed crops: Pinus koraiensis seed orchard as an example. Can. J. For. Res. 2017, 47, 366–370. [Google Scholar] [CrossRef]
- Kang, K.S.; Bilir, N. Seed Orchards (Establishment, Management and Genetics); OGEM-VAK Press: Ankara, Türkiye, 2021; Available online: https://www.ogemvak.org.tr/ (accessed on 10 March 2023).
- El-Kassaby, Y. Evaluation of the tree-improvement delivery system: Factors affecting genetic potential. Tree Physiol. 1995, 15, 545–550. [Google Scholar] [CrossRef]
- Eriksson, G.; Lindgren, D.; Jonsson, A. Flowering in a Clone Trial of Picea abies Karst; Technical Report; Royal School of Forestry: Stockholm, Sweden, 1973; Volume 110, pp. 1–45. [Google Scholar]
- Griffin, A.R. Clonal variation in radiata pine seed orchards. I. Some flowering, cone, and seed production traits. Aust. For. Res. 1982, 12, 295–302. Available online: http://jkv.50megs.com/afr.html (accessed on 29 April 2023).
- Roeder, K.; Devlin, B.; Lindsay, B.G. Application of maximum likelihood methods to population genetic data for the estimation of individual fertilities. Biometrics 1989, 45, 363–379. [Google Scholar] [CrossRef]
- El-Kassaby, Y.A.; Reynolds, S. Reproductive phenology, parental balance and supplemental mass pollination in a Sitka spruce seed orchard. For. Ecol. Manag. 1990, 31, 45–54. [Google Scholar] [CrossRef]
- Xie, C.Y.; Knowles, P. Male fertility variation in an open-pollinated plantation of Norway spruce (Picea abies). Can. J. For. Res. 1992, 22, 1463–1468. [Google Scholar] [CrossRef]
- El-Kassaby, Y.A.; Cook, C. Female reproductive energy and reproductive success in a Douglas-fir seed orchard and its impact on genetic diversity. Silvae Genet. 1994, 43, 243–246. [Google Scholar]
- Gömöry, D.; Bruchanik, R.; Paule, L. Effective population number estimation of three Scots pine (Pinus sylvestris L.) seed orchards based on an integrated assessment of flowering, floral phenology, and seed orchard design. For. Genet. 2000, 7, 65–75. [Google Scholar]
- Pakkanen, A.; Nikkanen, T.; Pulkkinen, P. Annual variation in pollen contamination and outcrossing in a Picea abies seed orchard. Scand. J. For. Res. 2000, 15, 399–404. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Kang, K.S.; Lindgren, D. Fertility variation among clones of Korean pine (Pinus koraiensis S. et Z.) and its implications on seed orchard management. For. Genet. 1999, 6, 191–200. Available online: https://kf.tuzvo.sk/sites/default/files/FG06-3_191-200.pdf (accessed on 20 April 2023).
- Bilir, N. Fertility variation in wild rose (Rosa canina) over habitat classes. Int. J. Agric. Biol. 2011, 13, 110–114. [Google Scholar]
- Burczyk, J.; Chalupka, W. Flowering, and cone production variability and its effect on parental balance in a Scots pine clonal seed orchard. Ann. For. Sci. 1997, 54, 129–144. Available online: https://hal.science/hal-00883136 (accessed on 10 March 2023). [CrossRef]
- Kang, K.S.; Lindgren, D. Fertility variation and its effect on the relatedness of seeds in Pinus densiflora, Pinus thunbergii and Pinus koraiensis clonal seed orchards. Silvae Genet. 1998, 47, 196–201. [Google Scholar]
- Bila, A.D.; Lindgren, D.; Mullin, T.J. Fertility variation and its effect on diversity over generations in a teak plantation (Tectona grandis L.f.). Silvae Genet. 1999, 48, 109–114. [Google Scholar]
- Bila, A.D.; Lindgren, D. Fertility variation in Milletias thuhlmannii, Brachystegia spiciformis, Brachystegia bohemii and Leucaena leucocephala and its effects on relatedness in seeds. For. Genet. 1998, 5, 119–129. Available online: https://kf.tuzvo.sk/sites/default/files/FG05-2_119-129.pdf (accessed on 21 May 2023).
- Kang, K.S.; El-Kassaby, Y.A. Considerations of correlated fertility between genders on genetic diversity: The Pinus densiflora seed orchard as a model. Theor. Appl. Genet. 2002, 105, 1183–1189. [Google Scholar] [CrossRef]
- Gömöry, D.; Bruchánik, R.; Longauer, R. Fertility variation, and flowering asynchrony in Pinus sylvestris: Consequences for the genetic structure of progeny in seed orchards. For. Ecol. Manag. 2003, 174, 117–126. Available online: https://www.sciencedirect.com/science/article/pii/S0378112702000312?via%3Dihub (accessed on 12 June 2023). [CrossRef]
- Varghese, M.; Lindgren, D.; Nicodemus, A. Fertility and effective population size in seedling seed orchards of Casuarina equisetifolia and C. junghuhniana. Silvae Genet. 2004, 53, 164–168. [Google Scholar] [CrossRef]
- Varghese, M.; Nicodemus, A.; Nagarajan, B.; Lindgren, D. Impact of fertility variation on gene diversity and drift in two clonal seed orchards of teak (Tectona grandis Linn. f.). New For. 2006, 31, 497–512. [Google Scholar] [CrossRef]
- Prescher, F.; Lindgren, D.; Almqvist, C.; Kroon, J.; Lestander, T.; Mullin, T.J. Female fertility variation in mature Pinus sylvestris clonal seed orchards. Scand. J. For. Res. 2007, 22, 280–289. [Google Scholar] [CrossRef]
- Kamalakannan, R.; Varghese, M.; Lindgren, D. Fertility Variation and its Implications on Relatedness in Seed Crops in Seedling Seed Orchards of Eucalyptus camaldulensis and E. tereticornis. Silvae Genet. 2007, 56, 253–259. [Google Scholar] [CrossRef]
- Ertekin, M. Clone fertility and genetic diversity in a Black pine seed orchard. Silvae Genet. 2010, 59, 145–150. [Google Scholar] [CrossRef][Green Version]
- Kamalakannan, R.; Varghese, M.; Suraj, P.G.; Arutselvan, T. Options for converting a clone trial of Eucalyptus camaldulensis into a clonal seed orchard considering gain, fertility and effective clone number. J. For. Res. 2016, 27, 51–57. [Google Scholar] [CrossRef]
- Cockerham, C.C. Group inbreeding and coancestry. Genetics 1967, 56, 89–104. [Google Scholar] [CrossRef]
- Kang, K.S.; Lindgren, D.; Mullin, T. Prediction of genetic gain and gene diversity in seed orchard crops under alternative management strategies. Theor. Appl. Genet. 2001, 103, 1099–1107. [Google Scholar] [CrossRef]
- Kartikawati, N. Fertility variation of Melaleuca cajuput subsp cajuputi and its implication seed orchard management. Indones. J. For. Res. 2016, 3, 83–94. [Google Scholar] [CrossRef]
- Kaya, Z.; Ozel, H.B. Fertility variation and gene diversity based on cone and seed production in a clonal seed orchard of Pinus nigra fresenius. Environ. Bull. 2018, 27, 3162–3165. Available online: https://www.prt-parlar.de/ (accessed on 11 March 2023).
- Suraj, P.G.; Nagabhushana, K.; Kamalakannan, R.; Varghese, M. Impact of fertility variation on genetic diversity and phenotypic traits in second generation seed production areas and clonal seed orchards of Eucalyptus camaldulensis. Silvae Genet. 2019, 68, 29–40. [Google Scholar] [CrossRef]
- Funda, T.; El-Kassaby, Y.A. Seed orchard genetics. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2012, 7, 13. [Google Scholar] [CrossRef]
- Liesebach, H.; Liepe, K.; Bäucker, C. Towards new seed orchard designs in Germany—A review. Silvae Genet. 2021, 70, 84–98. [Google Scholar] [CrossRef]
- Muñoz-Gutiérrez, L.; Vargas-Hernández, J.J.; López-Upton, J.; Ramírez-Herrera, C.; Jiménez-Casas, M. Clonal variation in phenological synchronization and cone production in a Pinus patula seed orchard. Silvae Genet. 2020, 69, 130–138. [Google Scholar] [CrossRef]
- Wu, H.X.; Ker, R.; Chen, Z.; Ivkovic, M. Balancing breeding for growth and fecundity in Radiata pine (Pinus radiata D. Don) breeding programme. Evol. Appl. 2021, 14, 834–846. [Google Scholar] [CrossRef]
- Galeta, P.; Pankowská, A. A new method for estimating growth and fertility rates using age-at-death ratios in small skeletal samples: The effect of mortality and stochastic variation. PLoS ONE 2023, 18, e0286580. [Google Scholar] [CrossRef] [PubMed]
- Muller-Starck, G.; Ziehe, M. Reproductive systems in conifer seed orchards. Theor. Appl. Genet. 1984, 69, 173–177. [Google Scholar] [CrossRef]
- Schmidtling, R.C. The inheritance of precocity and its relationship with growth in loblolly pine. Silvae Genet. 1981, 30, 188–192. [Google Scholar]
- Schultz, R.P. Stimulation of Flower and Seed Production in a Young Slash Pine Orchard; Southeastern Forest Experiment Station Forest Service, U.S. Department of Agriculture: New Orleans, LA, USA, 1971; 10p.
- Savolainen, O.; Karkkainen, K.; Harju, A.; Nikkanen, T.; Rusanen, M. Fertility variation in Pinus sylvestris: A test of sexual allocation theory. Am. J. Bot. 1993, 80, 1016–1020. [Google Scholar] [CrossRef]
- Hannerz, M.; Aitken, S.; Ericsson, N.; Ying, C.C. Inheritance of strobili production and genetic correlation with growth in lodgepole pine. For. Genet. 2001, 8, 323–329. [Google Scholar]
- Bhumibhamon, S. Studies on Scots pine seed orchards in Finland with special emphasis on the genetic composition of the seed. Comm. Inst. For. Fenn. 1978, 94, 1–118. [Google Scholar]
- Kjær, E.D.; Wellendorf, H. Variation in flowering and reproductive success in a Danish Picea abies (Karst) seed orchard. For. Genet. 1997, 4, 181–188. [Google Scholar]
- Nikkanen, T.; Ruotsalainen, S. Variation in flowering abundance and impact on the genetic diversity of the seed crop in a Norway Spruce seed orchard. Silva Fenn. 2000, 34, 205–222. [Google Scholar] [CrossRef]
- Nikkanen, T.; Velling, P. Correlations between flowering and some vegetative characteristics of grafts of Pinus sylvestris. For. Ecol. Manag. 1987, 19, 35–40. [Google Scholar] [CrossRef]
- Almqvist, C.; Jansson, G.; Sonesson, J. Genotypic correlations between early cone-set and height growth in Picea abies clonal trials. For. Genet. 2001, 8, 197–204. [Google Scholar]
- Boydak, M. Seed Yield of Pinus sylvestris in Catacik-Eskisehir; Istanbul University Press: Istanbul, Türkiye, 1977; Available online: www.istanbul.edu.tr (accessed on 11 May 2023).
- Lindgren, D.; Gea, L.; Jefferson, P. Loss of genetic diversity monitored by status number. Silvae Genet. 1996, 45, 52–59. Available online: https://www.degruyter.com (accessed on 12 June 2023).
- Ivetić, V.; Devetaković, J.; Nonić, M.; Stanković, D.; Šijačić-Nikolić, M. Genetic diversity and forest reproductive material—From seed source selection to planting. iForest 2016, 9, 801–812. [Google Scholar] [CrossRef]
- Matziris, D. Variation in cone production in a clonal seed orchard of Black pine. Silvae Genet. 1993, 42, 136–141. [Google Scholar]
- Park, J.M.; Kang, H.Y.; Yeom, D.B.; Kang, K.S.; El-Kassaby, Y.A.; Lee, K.M. Gender, reproductive output covariation and their role on gene diversity of Pinus koraiensis seed orchard crops. BMC Plant Biol. 2020, 20, 418. [Google Scholar] [CrossRef]
- Lloyd, D.G. Parental strategies of angiosperms. N. Z. J. Bot. 1979, 17, 595–606. [Google Scholar] [CrossRef]
- Jiao, S.-Q.; Li, M.; Zhu, Y.-J.; Zhou, S.-S.; Zhao, S.-W.; Li, Z.-C.; Bao, Y.-T.; Shi, T.-L.; Zhang, H.-J.; Yang, X.-L. Variation in Platycladus orientalis (Cupressaceae) Reproductive output and its effect on seed orchard crops’ genetic diversity. Forests 2021, 12, 1429. [Google Scholar] [CrossRef]
- Kang, K.S. Clonal and annual variation of flower production and composition of gamete gene pool in a clonal seed orchard of Pinus densiflora. Can. J. For. Res. 2000, 30, 1275–1280. [Google Scholar] [CrossRef]
- Eriksson, V.J.; Adams, W.T. Mating success in a coastal Douglas-fir seed orchard as affected by distance and floral phenology. Can. J. For. Res. 1989, 19, 1248–1255. [Google Scholar] [CrossRef]
- El Kassaby, Y.A.; Ritland, K.; Fashler, A.M.; Devitt, W.J.B. The role of reproductive phenology upon the mating success of a Douglas-fir seed orchard. Silvae Genet. 1988, 37, 76–82. [Google Scholar]
- Xie, J.; Huang, X.; Liu, Y.; Zhu, P.; Zhu, Y.; Li, F.; Yao, J.; Chen, L.; Yang, H. Variation of fertility and phenological synchronization in Cunninghamia lanceolata seed orchard: Implications for seed production. Forests 2022, 13, 1571. [Google Scholar] [CrossRef]
- Askew, G.R.; Blush, T.D. Short note: An index of phenological overlap in flowering for clonal conifer seed orchards. Silvae Genet. 1990, 39, 168–171. [Google Scholar]
- Alexander, L.W.; Woeste, K.E. Phenology, dichogamy, and floral synchronization in a northern red oak (Quercus rubra) seed orchard. Can. J. For. Res. 2016, 46, 629–636. [Google Scholar] [CrossRef]
- Mápula-larreta, M.; López-Upton, J.; Vargas-Hernández, J.J.; Hernández-Livera, A. Reproductive indicators in natural populations of Douglas-fir in Mexico. Biodivers. Conserv. 2007, 16, 727–742. [Google Scholar] [CrossRef]
- El-Kassaby, Y.A.; Funda, T.; Lai, B.S.K. Female reproductive success variation in a Pseudotsuga menziesii seed orchard as revealed by pedigree reconstruction from a bulk seed collection. J. Hered. 2010, 101, 164–168. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, K.-S.; Bilir, N.; Jeon, K.; Kim, Y.-J. Fertility Variation and Gene Diversity in Forest Populations. Forests 2023, 14, 2172. https://doi.org/10.3390/f14112172
Kang K-S, Bilir N, Jeon K, Kim Y-J. Fertility Variation and Gene Diversity in Forest Populations. Forests. 2023; 14(11):2172. https://doi.org/10.3390/f14112172
Chicago/Turabian StyleKang, Kyu-Suk, Nebi Bilir, Koeun Jeon, and Ye-Ji Kim. 2023. "Fertility Variation and Gene Diversity in Forest Populations" Forests 14, no. 11: 2172. https://doi.org/10.3390/f14112172
APA StyleKang, K.-S., Bilir, N., Jeon, K., & Kim, Y.-J. (2023). Fertility Variation and Gene Diversity in Forest Populations. Forests, 14(11), 2172. https://doi.org/10.3390/f14112172






