An Analysis of the Factors Affecting Forest Mortality and Research on Forecasting Models in Southern China: A Case Study in Zhejiang Province
Abstract
:1. Introduction
2. Study Areas and Methods
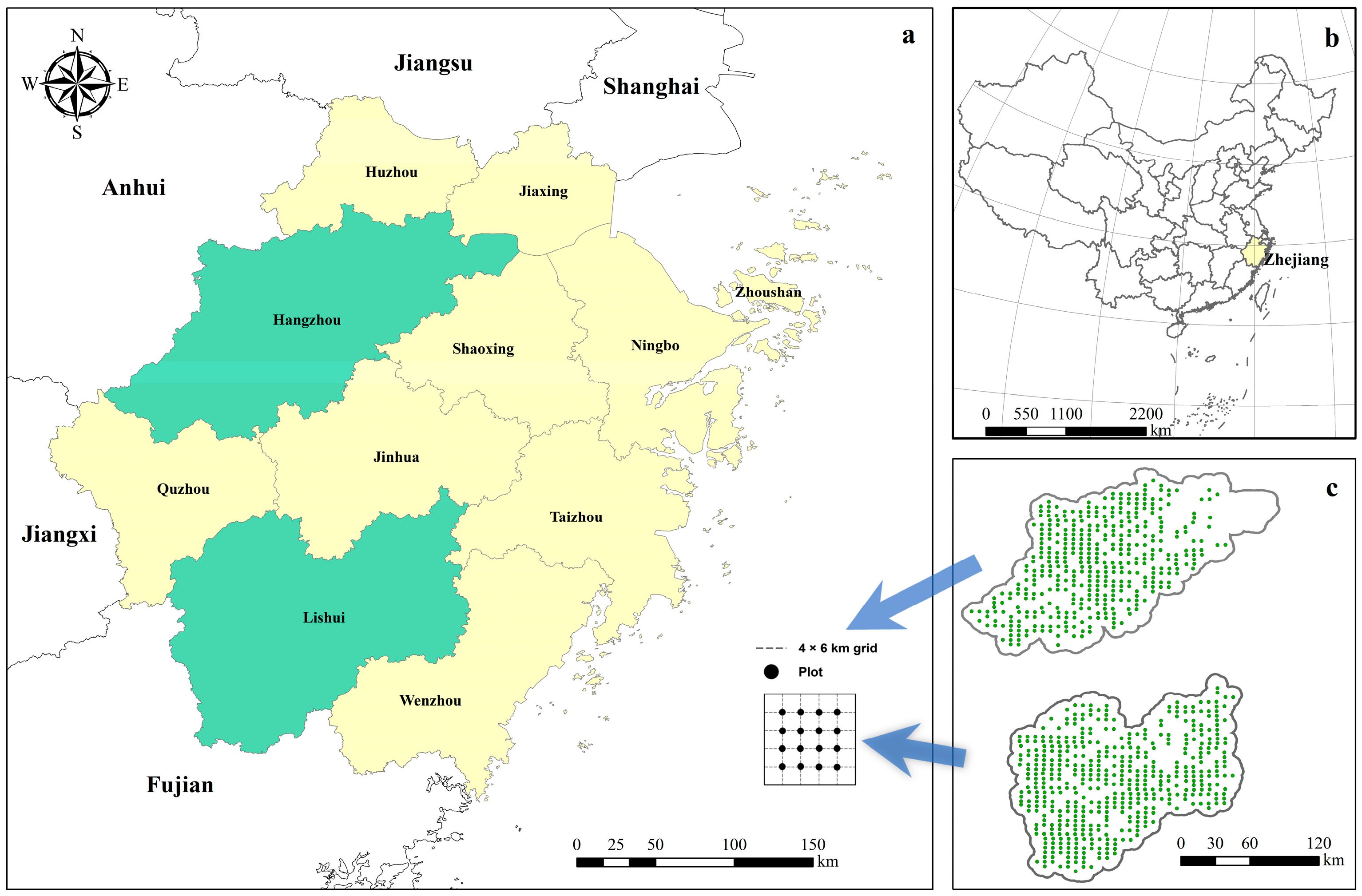
2.1. Description of Study Area
2.2. Selection of Predictor Variables
3. Models Selection and Approaches
3.1. Training Datasets
3.2. Linear Regression Models
3.3. Machine Learning Models
3.4. Data Structure
3.5. Model Evaluation
4. Results
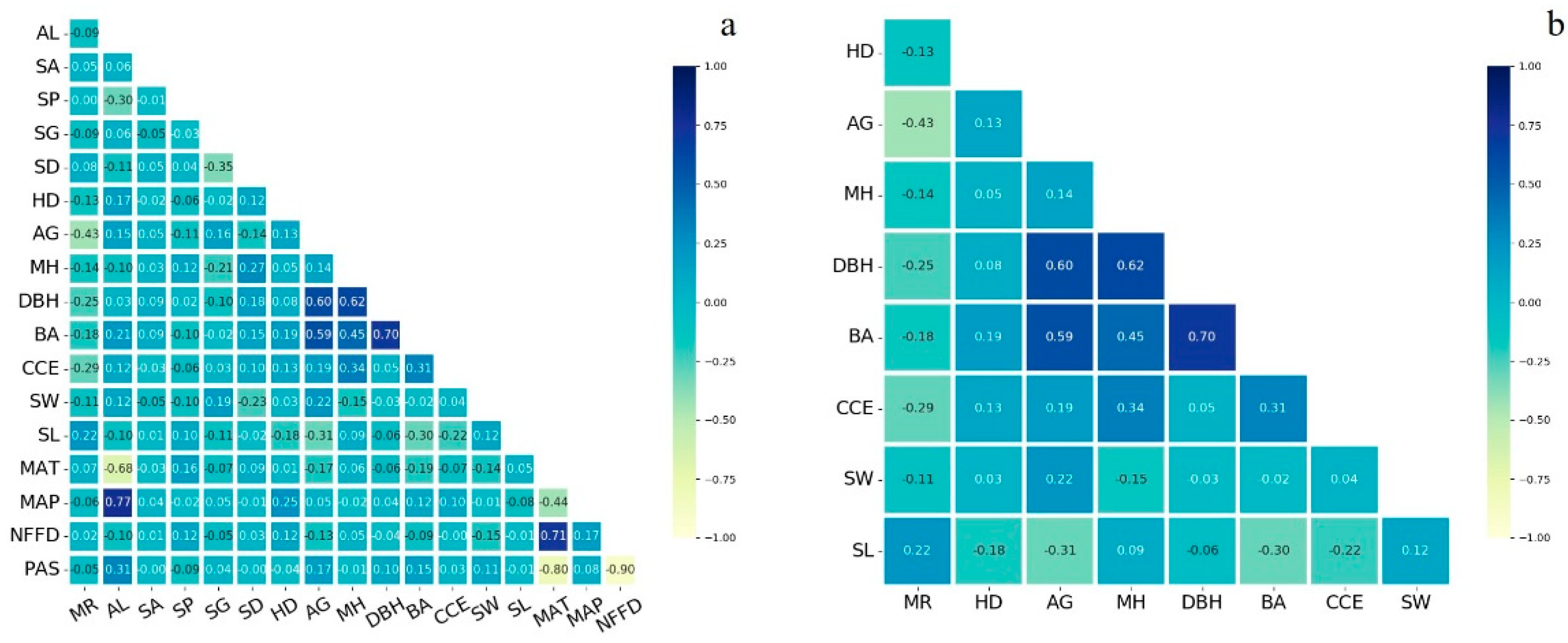
4.1. Linear Regression Modelling Results
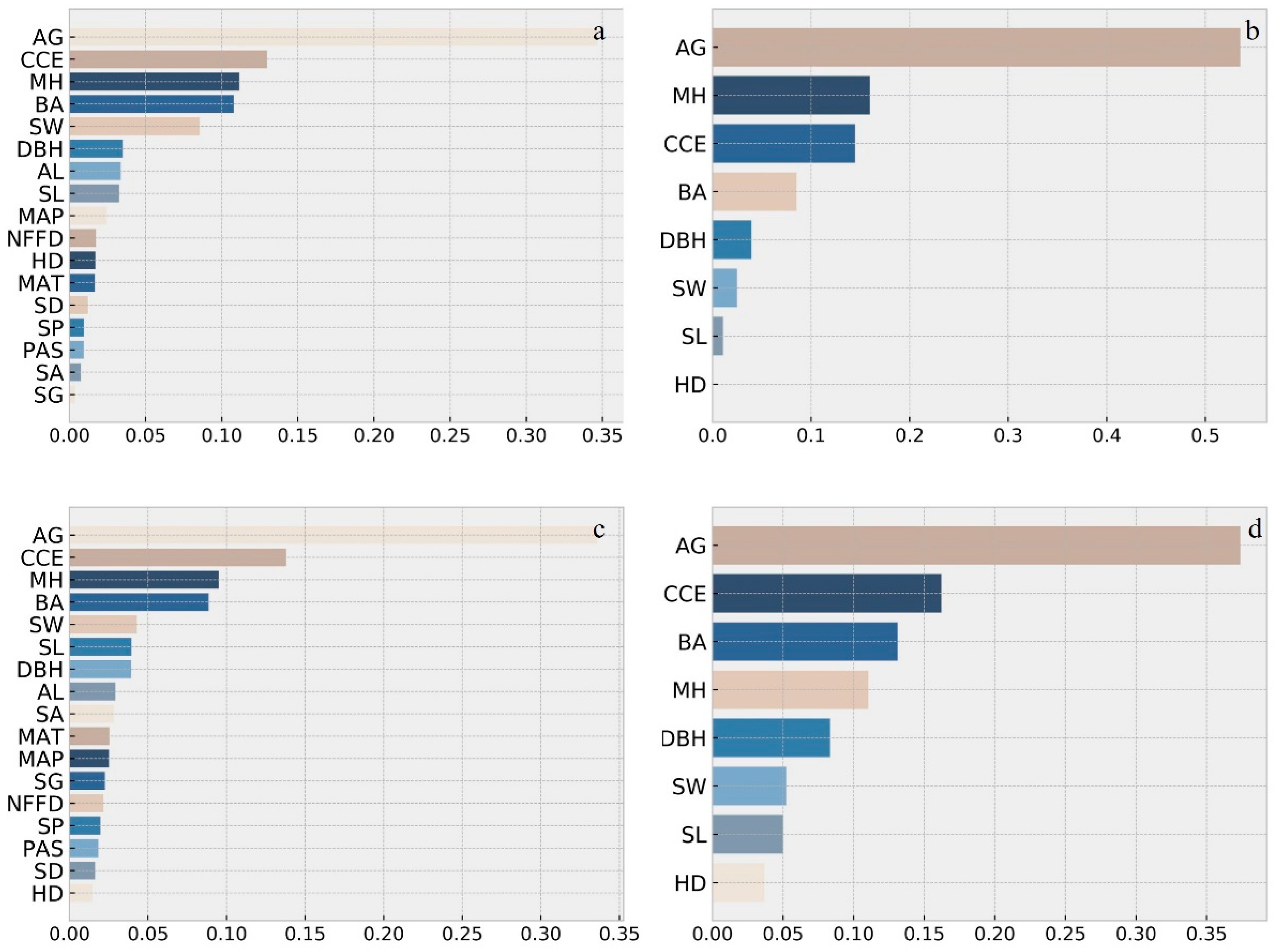
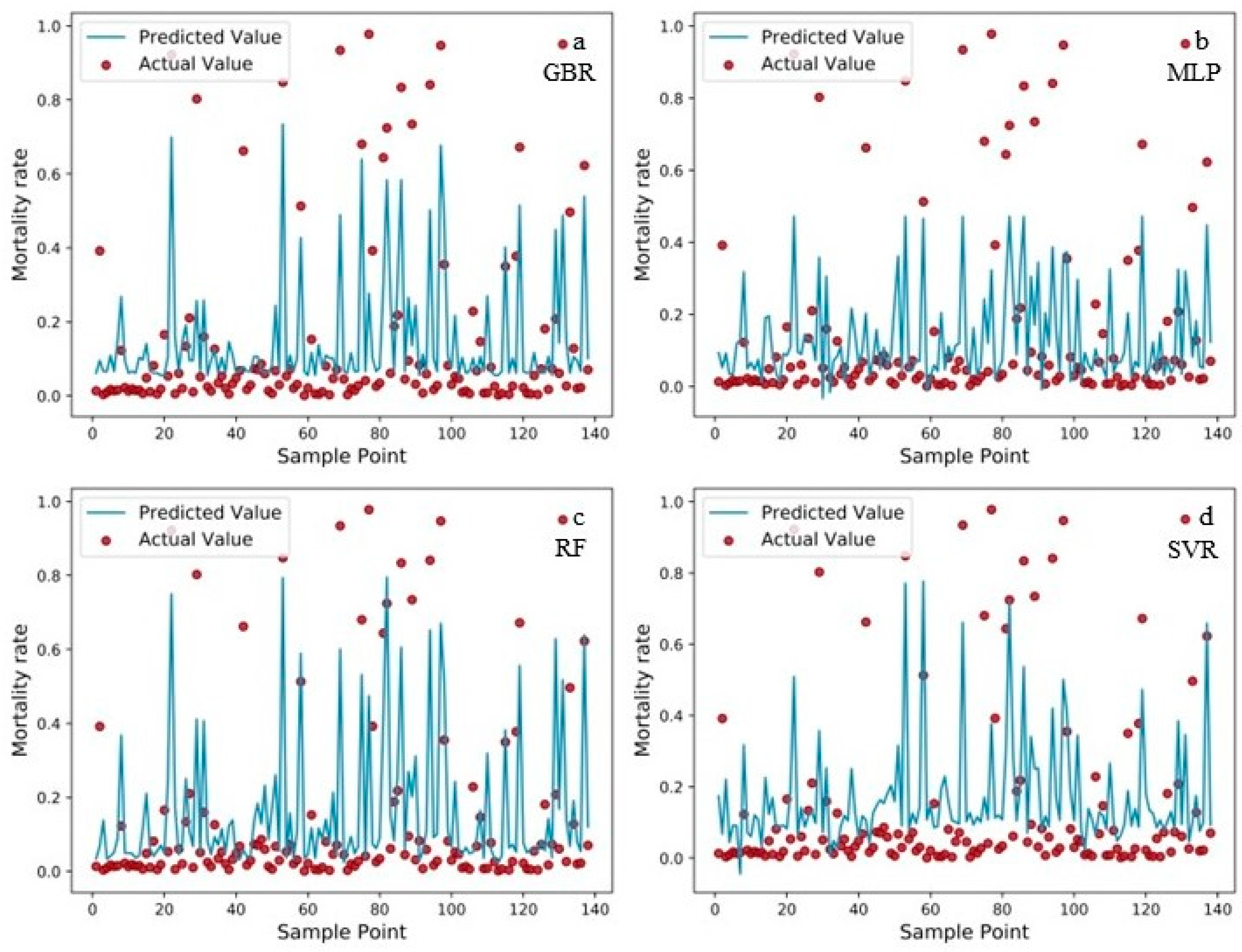
4.2. Machine Learning Modelling Results
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chambers, J.Q.; Higuchi, N.; Teixeira, L.M.; dos Santos, J.; Laurance, S.G.; Trumbore, S.E. Response of tree biomass and wood litter to disturbance in a Central Amazon forest. Oecologia 2004, 141, 596–611. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Peng, C.; Schneider, R.; Cyr, D.; McDowell, N.G.; Kneeshaw, D. Drought-induced increase in tree mortality and corresponding decrease in the carbon sink capacity of Canada’s boreal forests from 1970 to 2020. Glob. Chang. Biol. 2020, 29, 2274–2285. [Google Scholar] [CrossRef]
- Perez-Quezada, J.F.; Barichivich, J.; Urrutia-Jalabert, R.; Carrasco, E.; Aguilera, D.; Bacour, C.; Lara, A. Warming and Drought Weaken the Carbon Sink Capacity of an Endangered Paleoendemic Temperate Rainforest in South America. J. Geophys. Res. Biogeosci. 2023, 128, e2022JG007258. [Google Scholar] [CrossRef] [PubMed]
- Eid, T.; Tuhus, E. Models for individual tree mortality in Norway. For. Ecol. Manag. 2001, 154, 69–84. [Google Scholar] [CrossRef]
- Xie, L.; Chen, X.; Zhou, X.; Sharma, R.P.; Li, J. Developing Tree Mortality Models Using Bayesian Modeling Approach. Forests 2022, 13, 604. [Google Scholar] [CrossRef]
- Yao, X.; Titus, S.J.; Macdonald, S.E. A generalized logistic model of individual tree mortality for aspen, white spruce, and lodgepole pine in Alberta mixed wood forests. Can. J. For. Res. 2001, 31, 283–291. [Google Scholar]
- Cailleret, M.; Bircher, N.; Hartig, F.; Hülsmann, L.; Bugmann, H. Bayesian calibration of a growth-dependent tree mortality model to simulate the dynamics of European temperate forests. Ecol. Appl. Publ. Ecol. Soc. Am. 2020, 30, e02021. [Google Scholar] [CrossRef]
- Batllori, E.; Lloret, F.; Aakala, T.; Anderegg, W.R.L.; Aynekulu, E.; Bendixsen, D.P.; Bentouati, A.; Bigler, C.; Burk, C.J.; Camarero, J.J.; et al. Forest and woodland replacement patterns following drought-related mortality. Proc. Natl. Acad. Sci. USA 2020, 117, 29720–29729. [Google Scholar] [CrossRef]
- Lun, F.; Liu, Y.; He, L.; Yang, L.; Liu, M.; Li, W. Life cycle research on the carbon budget of the Larix principis-rupprechtii plantation forest ecosystem in North China. J. Clean. Prod. 2018, 177, 178–186. [Google Scholar] [CrossRef]
- Zhao, D.; Borders, B.; Wilson, M. Individual-tree diameter growth and mortality models for bottomland mixed-species hardwood stands in the lower Mississippi alluvial valley. For. Ecol. Manag. 2004, 199, 307–322. [Google Scholar] [CrossRef]
- Li, R.; Weiskittel, A.R.; John, A.; Kershaw, J. Modeling annualized occurrence, frequency, and composition of ingrowth using mixed-effects zero-inflated models and permanent plots in the Acadian Forest Region of North America. Can. J. For. Res. 2011, 41, 2077–2089. [Google Scholar] [CrossRef]
- Qiu, S.; Xu, M.; Li, R.; Zheng, Y.; Clark, D.; Cui, X.; Liu, L.; Lai, C.; Zhang, W.; Liu, B. Climatic information improves statistical individual-tree mortality models for three key species of Sichuan Province, China. Ann. For. Sci. 2015, 72, 443–455. [Google Scholar] [CrossRef]
- Salas-Eljatib, C.; Weiskittel, A.R. On studying the patterns of individual-based tree mortality in natural forests: A modelling analysis. For. Ecol. Manag. 2020, 475, 118369. [Google Scholar] [CrossRef]
- Zhou, X.; Fu, L.; Sharma, R.P.; He, P.; Lei, Y.; Guo, J. Generalized or general mixed-effect modelling of tree morality of Larix gmelinii subsp. principis-rupprechtii in Northern China. J. For. Res. 2021, 32, 2447–2458. [Google Scholar] [CrossRef]
- Zhang, X.; Lei, Y.; Lei, X.; Chen, Y.; Feng, M. Predicting Stand-Level Mortality with Count Data Models. Sci. Silvae Sin. 2012, 48, 54–61. (In Chinese) [Google Scholar]
- Hu, M.-C.; Pavlicova, M.; Nunes, E.V. Zero-Inflated and Hurdle Models of Count Data with Extra Zeros: Examples from an HIV-Risk Reduction Intervention Trial. Am. J. Drug Alcohol Abus. 2011, 37, 367–375. [Google Scholar] [CrossRef]
- Feng, C.X. A comparison of zero-inflated and hurdle models for modeling zero-inflated count data. Feng J. Stat. Distrib. Appl. 2021, 8, 8. [Google Scholar] [CrossRef]
- Bircher, N.; Cailleret, M.; Bugmann, H. The agony of choice: Different empirical mortality models lead to sharply different future forest dynamics. Ecol. Appl. 2015, 25, 1303–1318. [Google Scholar] [CrossRef]
- Guan, H.; Dong, X.; Yan, G.; Searls, T.; Bourque, C.P.A.; Meng, F.R. Conditional inference trees in the assessment of tree mortality rates in the transitional mixed forests of Atlantic Canada. PLoS ONE 2021, 16, e0250991. [Google Scholar] [CrossRef]
- Hülsmann, L.; Bugmann, H.; Cailleret, M.; Brang, P. How to Kill a Tree: Empirical Mortality Models for 18 Species and Their Performance in a Dynamic Forest Model. Ecol. Appl. 2018, 28, 522–540. [Google Scholar] [CrossRef]
- McNellis, B.E.; Smith, A.M.S.; Smith, A.T.; Strand, E.K. Tree mortality in western U.S. forests forecasted using forest inventory and Random Forest classification. Ecosphere 2021, 12, e03419. [Google Scholar] [CrossRef]
- Ma, W.; Lin, G.; Liang, J. Estimating dynamics of central hardwood forests using random forests. Ecol. Model. 2020, 419, 108947. [Google Scholar] [CrossRef]
- Moore, J.A.; Hamilton, D.A., Jr.; Xiao, Y.; Byrne, J. Bedrock type significantly affects individual tree mortality for various conifers in the inland Northwest, U.S.A. Can. J. For. Res. 2004, 34, 31–42. [Google Scholar] [CrossRef]
- Wang, W.; Peng, C.; Kneeshaw, D.D.; Larocque, G.R.; Luo, Z. Drought-induced tree mortality: Ecological consequences, causes, and modeling. Environ. Rev. 2012, 20, 109–121. [Google Scholar] [CrossRef]
- Das, A.J.; Stephenson, N.L. Improving estimates of tree mortality probability using potential growth rate. Can. J. For. Res. 2015, 45, 920–928. [Google Scholar] [CrossRef]
- Hurst, J.M.; Stewart, G.H.; Perry, G.L.W.; Wiser, S.K.; Norton, D.A. Determinants of tree mortality in mixed old-growth Nothofagus forest. For. Ecol. Manag. 2012, 270, 189–199. [Google Scholar] [CrossRef]
- Timilsina, N.; Staudhammer, C.L. Individual Tree Mortality Model for Slash Pine in Florida: A Mixed Modeling Approach. South. J. Appl. For. 2012, 36, 211–219. [Google Scholar] [CrossRef]
- Vanoni, M.; Bugmann, H.; Nötzli, M.; Bigler, C. Drought and frost contribute to abrupt growth decreases before tree mortality in nine temperate tree species. For. Ecol. Manag. 2016, 382, 51–63. [Google Scholar] [CrossRef]
- Yaussy, D.A.; Iverson, L.R.; Matthews, S.N. Competition and Climate Affects US Hardwood-Forest Tree Mortality. For. Sci. 2013, 59, 416–430. [Google Scholar] [CrossRef]
- Shao, F.; Yu, X.; Zheng, J.; Wang, H. Relationships between dominant arbor species distribution and environmental factors of shelter forests in the Beijing mountain area. Acta Ecol. Sin. 2012, 32, 6092–6099. (In Chinese) [Google Scholar] [CrossRef]
- Wang, T.; Wang, G.; Innes, J.L.; Seely, B.; Chen, B. ClimateAP: An application for dynamic local downscaling of historical and future climate data in Asia Pacific. Front. Agric. Sci. Eng. 2017, 4, 448–458. [Google Scholar] [CrossRef]
- Zhou, P.; Zhang, L.; Qi, S. Plant Diversity and Aboveground Biomass Interact with Abiotic Factors to Drive Soil Organic Carbon in Beijing Mountainous Areas. Sustainability 2022, 14, 10655. [Google Scholar] [CrossRef]
- Liu, X.; Lin, L.; Wang, Y. Improving the multiple linear regression method of biomass estimation using plant water-based spectrum correction. Remote Sens. Lett. 2022, 13, 716–725. [Google Scholar] [CrossRef]
- Jiménez, R.D.L.; Gao, Y.; Solórzano, J.V.; Skutsch, M.; Pérez SD, R.; Salinas, M.M.A.; Farfán, M. Mapping Forest Degradation and Contributing Factors in a Tropical Dry Forest. Front. Environ. Sci. 2022, 10, 912873. [Google Scholar] [CrossRef]
- Liu, C.; Chen, D.; Zou, C.; Liu, S.; Li, H.; Liu, Z.; Ye, L. Modeling Biomass for Natural Subtropical Secondary Forest Using Multi-Source Data and Different Regression Models in Huangfu Mountain, China. Sustainability 2022, 14, 13006. [Google Scholar] [CrossRef]
- Wohlgemuth, T. Modelling floristic species richness on a regional scale: A case study in Switzerland. Biodivers. Conserv. 1998, 7, 159–177. [Google Scholar] [CrossRef]
- Cai, T.; Ju, C.; Yang, X. Comparison of Ridge Regression and Partial Least Squares Regression for Estimating Above-Ground Biomass with Landsat Images and Terrain Data in Mu Us Sandy Land, China. Arid. Land Res. Manag. 2009, 23, 248–261. [Google Scholar] [CrossRef]
- Ohsowski, B.M.; Dunfield, K.E.; Klironomos, J.N.; Hart, M.M. Improving plant biomass estimation in the field using partial least squares regression and ridge regression. Botany 2016, 94, 501–508. [Google Scholar] [CrossRef]
- Bai, H.; Li, L.; Wu, Y.; Feng, G.; Gong, Z.; Sun, G. Identifying Critical Meteorological Elements for Vegetation Coverage Change in China. Front. Phys. 2022, 10, 3. [Google Scholar] [CrossRef]
- Kaźmierczak, K.; Zawieja, B. The influence of weather conditions on annual height increments of Scots pine. Biom. Lett. 2014, 51, 143–152. [Google Scholar] [CrossRef]
- Liu, J.; Pan, P.; Guo, Y.; Zang, H.; Ouyang, X.; You, J. Stand-Level Mortality Model of Cunninghamia lanceolata Forest in Southern Jiangxi Based on Zero-Inflated Model and Hurdle Model. Acta Agric. Univ. Jiangxiensis 2022, 44, 1428–1437. (In Chinese) [Google Scholar]
- Cai, J.; Xu, K.; Zhu, Y.; Hu, F.; Li, L. Prediction and analysis of net ecosystem carbon exchange based on gradient boosting regression and random forest. Appl. Energy 2020, 262, 114566. [Google Scholar] [CrossRef]
- Chen, M.; Qiu, X.; Zeng, W.; Peng, D. Combining Sample Plot Stratification and Machine Learning Algorithms to Improve Forest Aboveground Carbon Density Estimation in Northeast China Using Airborne LiDAR Data. Remote Sens. 2022, 14, 1477. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Spinger: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Zhou, Z.H. Machine Learning; Tsinghua University Press: Beijing, China, 2016. [Google Scholar]
- Ding, Y.; Zhang, H.; Wang, Z.; Xie, Q.; Wang, Y.; Liu, L.; Hall, C.C. A Comparison of Estimating Crop Residue Cover from Sentinel-2 Data Using Empirical Regressions and Machine Learning Methods. Remote Sens. 2020, 12, 1470. [Google Scholar] [CrossRef]
- Sohn, I.; Shim, J.; Hwang, C.; Kim, S.; Lee, J.W. Informative transcription factor selection using support vector machine-based generalized approximate cross validation criteria. Comput. Stat. Data Anal. 2008, 53, 1727–1735. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. J. Mach. Learn. 2010, 9, 249–256. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 11–18 December 2015; pp. 1026–1034. [Google Scholar]
- Akaike, H. A Bayesian analysis of the minimum AIC procedure. Ann. Inst. Stat. Math. 1978, 30, 9–14. [Google Scholar] [CrossRef]
- Chen, S.S.; Gopalakrishnan, P.S. Clustering via the Bayesian information criterion with applications in speech recognition. In Proceedings of the 1998 IEEE International Conference on Acoustics, Speech and Signal Processing, ICASSP ’98 (Cat. No.98CH36181), Seattle, WA, USA, 15 May 1998; IEEE: New York, NY, USA, 1998; Volume 2, pp. 645–648. [Google Scholar]
- Mengku, Z.; Lichun, J. Prediction of bark thickness for Larix gmelinii based on machine learning. J. Beijing For. Univ. 2022, 44, 54–62. (In Chinese) [Google Scholar]
- Wen, B.; Dong, W.; Xie, W.; Jun, M.A. Parameter optimization method for random forest based on improved grid search algorithm. Comput. Eng. Appl. 2018, 54, 154–157. (In Chinese) [Google Scholar]
- Calama, R.; Montero, G. Interregional nonlinear height–diameter model with random coefficients for stone pine in Spain. Can. J. For. Res. 2004, 34, 150–163. [Google Scholar] [CrossRef]
- Zanella, L.; Folkard, A.M.; Blackburn, G.A.; Carvalho, L.M.T. How well does random forest analysis model deforestation and forest fragmentation in the Brazilian Atlantic forest? Environ. Ecol. Stat. 2017, 24, 529–549. [Google Scholar] [CrossRef]
- Stephenson, N.L.; van Mantgem, P.J.; Bunn, A.G.; Bruner, H.; Harmon, M.E.; O’Connell, K.B.; Urban, D.L.; Franklin, J.F. Causes and implications of the correlation between forest productivity and tree mortality rates. Ecol. Monogr. 2011, 81, 527–555. [Google Scholar] [CrossRef]
- Wu, H.; Franklin, S.B.; Liu, J.; Lu, Z. Relative importance of density dependence and topography on tree mortality in a subtropical mountain forest. For. Ecol. Manag. 2017, 384, 169–179. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Yang, X.; Liu, L.; Li, X. Mortality models of semi-natural larch-spruce-fir (Larix olgensis-Picea jezoensis-Abies nephrolepis) forests based on soil factors. J. Fujian Agric. For. Univ. Nat. Sci. Ed. 2015, 44, 378–383. (In Chinese) [Google Scholar]
- Wang, T.; Dong, L.; Li, F. Laws and models of stand trees mortality for Hybrid larch young plantation. J. Northeast. For. Univ. 2017, 45, 39–43;48. (In Chinese) [Google Scholar]
- Caspersen, J.P. Variation in stand mortality related to successional composition. For. Ecol. Manag. 2004, 200, 149–160. [Google Scholar] [CrossRef]
- Zhao, D.; Borders, B.; Wang, M.; Kane, M. Modeling mortality of second-rotation loblolly pine plantations in the Piedmont/Upper Coastal Plain and Lower Coastal Plain of the southern United States. For. Ecol. Manag. 2007, 252, 132–143. [Google Scholar] [CrossRef]
- Hallinger, M.; Johansson, V.; Schmalholz, M.; Sjöberg, S.; Ranius, T. Factors driving tree mortality in retained forest fragments. For. Ecol. Manag. 2016, 368, 163–172. [Google Scholar] [CrossRef]
- Palmas, S.; Moreno, P.C.; Cropper, W.P.; Ortega, A.; Gezan, S.A. Stand-Level Components of a Growth and Yield Model for Nothofagus Mixed Forests from Southern Chile. Forests 2020, 11, 810. [Google Scholar] [CrossRef]
- Grodzki, W.; Oszako, T. Current Problems of Forest Protection in Spruce Stands under Conversion; Forest Research Institute: Warsaw, Poland, 2006. [Google Scholar]
- Breshears, D.D.; Myers, O.B.; Meyer, C.W.; Barnes, F.J.; Zou, C.B.; Allen, C.D.; Pockman, W.T. Tree die-off in response to global-change type drought: Mortality insights from a decade of plant water potential measurements. Front. Ecol. Environ. 2009, 7, 185–189. [Google Scholar] [CrossRef]
- Tyburski, Ł.; Zaniewski, P.T.; Bolibok, L.; Piątkowski, M.; Szczepkowski, A. Scots pine Pinus sylvestris mortality after surface fire in oligotrophic pine forest Peucedano-Pinetum in Kampinos National Park. Folia For. Pol. Ser. A For. 2019, 61, 51–57. [Google Scholar] [CrossRef]
- Zhang, X.; Lei, Y.; Pang, Y.; Liu, X.; Wang, J. Tree mortality in response to climate change induced drought across Beijing, China. Clim. Chang. 2014, 124, 179–190. [Google Scholar] [CrossRef]
- Dudek, T.; Grużewska, A. The type and extent of damages made by abiotic and biotic factors in managed forests of North−Eastern Poland. Sylwan 2022, 166, 41–53. [Google Scholar]
- Kautz, M.; Meddens, A.J.H.; Hall, R.J.; Arneth, A. Biotic disturbances in Northern Hemisphere forests—A synthesis of recent data, uncertainties and implications for forest monitoring and modelling. Glob. Ecol. Biogeogr. 2016, 26, 533–552. [Google Scholar] [CrossRef]
- Sierota, Z.; Grodzki, W.; Szczepkowski, A. Abiotic and Biotic Disturbances Affecting Forest Health in Poland over the Past 30 Years: Impacts of Climate and Forest Management. Forests 2019, 10, 75. [Google Scholar] [CrossRef]
- Bytnerowicz, A.; Szaro, R.; Karnosky, D.; Manning, W.; McManus, M.; Musselman, R.; Muzika, R.M. Importance of international research cooperative programs for better understanding of air pollution effects on forest ecosystems in Central Europe. In Effects of Air Pollution on Forest Health and Biodiversity in Forests of the Carpathian Mountains, Proceedings of the NATO Advanced Research Workshop, Stara Lesna, Slovakia, 22–26 May 2002; IOS Press: Amsterdam, The Netherlands, 2002; Volume 345, pp. 13–20. [Google Scholar]


| Scales | Variables | Min | Max | Mean | Standard Error |
|---|---|---|---|---|---|
| Terrain variables (T) | AL (m) | 6.00 | 1550.00 | 571.53 | 11.83 |
| SA | Qualitative variable | ||||
| SP | Qualitative variable | ||||
| SG (°) | 0.00 | 55.00 | 32.95 | 0.30 | |
| Ground variables (G) | SD (cm) | 20.00 | 120.00 | 46.52 | 0.49 |
| HD (cm) | 0.00 | 15.00 | 5.08 | 0.09 | |
| Stand variables (S) | AG (a) | 1.00 | 60.00 | 26.12 | 0.45 |
| DH (m) | 1.50 | 18.70 | 9.48 | 0.10 | |
| ADBH (cm) | 1.00 | 35.70 | 11.62 | 0.15 | |
| BA (m2·ha−1) | 0.01 | 240.83 | 37.34 | 1.31 | |
| CCE | 20.00 | 94.00 | 67.41 | 0.60 | |
| SW | 0.00 | 2.89 | 1.61 | 0.02 | |
| SI | 0.13 | 1.00 | 0.50 | 0.01 | |
| Climate variables (C) | MAT (°C) | 13.04 | 19.05 | 17.07 | 0.04 |
| MAP (mm) | 1421.45 | 2526.00 | 1836.92 | 8.99 | |
| NFFD (d) | 288.27 | 353.55 | 338.68 | 0.35 | |
| PAS (mm) | 1.45 | 37.73 | 5.81 | 0.15 | |
| Evaluation Index | M1 | M2 | M3 | M4 |
|---|---|---|---|---|
| AIC | −257.1192 | −272.1670 | 823.9227 | −4.8854 |
| BIC | −179.5080 | −237.6731 | 892.9105 | 95.0836 |
| −2 logL | 0.0344 | 0.0347 | 2355.5930 | 18.9521 |
| Evaluation Index | M1 | M2 | M3 | M4 |
|---|---|---|---|---|
| R² | 0.2791 | 0.2589 | 0.0733 | 0.2484 |
| MSE | 0.0405 | 0.0416 | 0.0495 | 0.0406 |
| MAE | 0.1341 | 0.1375 | 0.1448 | 0.1343 |
| EVS | 0.2791 | 0.2589 | 0.1502 | 0.2826 |
| RMSE | 0.2012 | 0.2040 | 0.2225 | 0.2015 |
| Model | Learning Rate | Max Depth | Min Samples Split | Number of Estimators |
|---|---|---|---|---|
| GBR | 0.16 | 2 | 5 | 20 |
| GBR_x17 | 0.10 | 3 | 7 | 100 |
| RF | 9 | 3 | 23 | |
| RF_x17 | 9 | 3 | 258 |
| Model | Learning Rate | Regularization Parameter (C) | Epsilon | Kernel Function Type | Hidden Layer Size | Maximum Number of Iterations |
|---|---|---|---|---|---|---|
| SVR | 1.00 | 0.10 | rbf | |||
| MLP | 0.01 | 0.10 | (16, 16) | 500 |
| R² | MSE | EVS | MAE | Accuracy | |
|---|---|---|---|---|---|
| GBR | 0.5912 | 0.0266 | 0.5915 | 0.1052 | 0.5912 |
| GBR_x17 | 0.6198 | 0.0247 | 0.6202 | 0.1015 | 0.6198 |
| RF | 0.6307 | 0.0240 | 0.6316 | 0.1032 | 0.6307 |
| RF_x17 | 0.6154 | 0.0250 | 0.6161 | 0.1046 | 0.6154 |
| SVR | 0.4949 | 0.0329 | 0.4984 | 0.1299 | 0.4949 |
| MLP | 0.4476 | 0.0359 | 0.4534 | 0.1236 | 0.4476 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Z.; Ji, B.; Yao, H.; Cheng, X.; Yu, S.; Sun, X.; Liu, S.; Xu, L.; Zhou, Y.; Shi, Y. An Analysis of the Factors Affecting Forest Mortality and Research on Forecasting Models in Southern China: A Case Study in Zhejiang Province. Forests 2023, 14, 2199. https://doi.org/10.3390/f14112199
Ding Z, Ji B, Yao H, Cheng X, Yu S, Sun X, Liu S, Xu L, Zhou Y, Shi Y. An Analysis of the Factors Affecting Forest Mortality and Research on Forecasting Models in Southern China: A Case Study in Zhejiang Province. Forests. 2023; 14(11):2199. https://doi.org/10.3390/f14112199
Chicago/Turabian StyleDing, Zhentian, Biyong Ji, Hongwen Yao, Xuekun Cheng, Shuhong Yu, Xiaobo Sun, Shuhan Liu, Lin Xu, Yufeng Zhou, and Yongjun Shi. 2023. "An Analysis of the Factors Affecting Forest Mortality and Research on Forecasting Models in Southern China: A Case Study in Zhejiang Province" Forests 14, no. 11: 2199. https://doi.org/10.3390/f14112199




