Abstract
Salt is a severe environmental stressor that affects growth and development in plants. It is significant to enhance the salt tolerance in plants. In this study, a salt-responsive WRKY transcription factor PtrWRKY51 was isolated from Populus trichocarpa (clone ‘Nisqually-1′). PtrWRKY51 was highly expressed in mature leaves and root and induced by salt stress. The PtrWRKY51 was overexpressed in Arabidopsis to investigate its biological functions. Compared with Col-0 lines, Overexpressed lines had an increase in germination rate of seed, root length, higher photosynthetic rate, instantaneous leaf WUE, chlorophyll content to improve salt tolerance under salt stress conditions. In contrast, compared to overexpressed and Col-0 lines, the mutant wrky51 was more sensitive to salt stress with lower photosynthetic rate and WUE. Additionally, it was found that the complementary lines (wrky51/ PtrWRKY51) had almost the same salt response as Col-0. In conclusion, PtrWRKY51 is a potential target in the enhancement of poplar tolerance by genetic engineering strategies.
1. Introduction
As one of the most widely distributed and adaptable forest species around the world, poplar is one of the most promising tree species for traditional afforestation and dealing with wood shortages. With the gradual aggravation of soil salinization, salt stress has become an essential factor restricting tree growth. Salt is one of the key environmental stress factors, which affects plant growth and development [1]. It is reported that more than 800 million hectares of land are affected by salinization in world, which across all continents including Africa, Asia, Australasia, Americas, and China, occupying an estimated 37.4 million hectares of salinized soil [2,3]. Moreover, soil salinization is becoming more serious due to environmental degradation, improper irrigation, climate change, growing population and industrial pollution [4,5]. Excessive salinity in plants will lead to excessive accumulation of intracellular metal ions, destroy ion balance, damage plant cell membrane structure, and thus affect the physiological and biochemical metabolic processes in plants [6,7]. Many studies have shown that the water potential of the soil in the saline-alkali soil was decreased, and the absorption of water by seed was inhibited, which seriously affected the germination rate, vigor index and germination index of seeds, which have a reduction in seed emergence rate and strong seedling rate [8,9]. Photosynthesis is the sum of a series of complex metabolic reactions which are essential in plant growth. Photosynthesis provides material and energy to maintain normal growth and development in plants, and chloroplasts in leaves are the place in which photosynthesis is carried out in higher plants. Salt stress affects plant absorb water and inorganic ions, inhibit the normal synthesis of chlorophyll, and reduce the content of chlorophyll, which affect the normal function of the pigment protein complexes, reduce the rate of plant convert light energy into chemical energy, which eventually make plant to a serious shortage of energy supply, inhibit the growth and development in plants [10,11]. At the same time, salt stress can also cause damage to chloroplast structure, and then affect its photosynthetic performance. It has been reported that under salt stress, the net photosynthetic rate, chlorophyll fluorescence and stomatal conductance of leaves are greatly reduced in citrus, which inhibit growth [12]. Similarly, it has been confirmed that salt stress seriously inhibits the photosynthetic rate and electron transport rate in photosynthesis, which affected plant growth in mustard [13]. Therefore, it is important to improve the salt tolerance in plants. During evolution, plants have developed various strategies to adapt to salt stress [14]. Signal sensing and signal transduction are key ways for plants to cope with challenges in environment, which involve many regulatory genes and proteins. Among these, the transcription factors, such as WRKY, NAC, bZIP, MYB, HSFs, AP2/EREBP have been identified as key regulators in controlling intrinsic development processes and regulating stress resistance by stimulating the expression of downstream targets in improving salt resistance in plants [15,16,17,18,19].
WRKY transcription factor is a key regulator in plants, and significant study has been made in WRKY transcription factor over the past 20 years [20]. SPF1 is the first WRKY gene, cloned from sweet potato [21]. Subsequently, WRKY genes were cloned and reported in Arabidopsis thaliana, wild oats and parsley, which were found that the proteins were encoded by WRKY genes that could specifically bind to DNA sequence element (T) (T) TGAC (C/T) [22]. The specific DNA sequence element (T) (T) TGAC (C/T) is also known as the W-box [22,23,24]. It is well known that WRKY transcription factor contain a highly conserved WRKY domains with WRKYGQK sequence and a zinc-finger-like motif CX4–7-CX23–28-HX1–2-(H/C) (C2H2 or C2HC) at N and C termini [25]. In general, WRKY proteins can be categorized into three groups based on the number of WRKY domains and the pattern of the zinc finger motif [25]. Group I WRKY transcription contains two WDs and C2H2 zinc finger, and group II and III WRKY transcription factors contain one WD and C2H2 zinc finger motif [25].
Many studies have found that WRKY transcription factors is a key role in plant response to abiotic stresses such as drought and salt stress. Northern blot hybridization showed that 10 WRKY genes in rice had different responses to salt, osmotic stress, cold and heat treatments [26]. In wheat, eight of the 15 WRKY genes also showed response to PEG, low temperature, high temperature and NaCl stress treatments [27]. In Arabidopsis, overexpression of OSWRKY45 and AtWRKY46 can regulate stomatal movement in response to drought and salt stresses [28,29]. Heterologous expression of GmWRKY54 in Arabidopsis increased drought and salt tolerance [30]. In addition, WRKY genes such as TaWRKY2 and TaWRKY19, TaWRKY146, TaWRKY1 and TaWRKY33, VaWRKY14, ZmWRKY17 and ZmWRKY40, and PeWRKY83 were overexpressed in Arabidopsis can improve plant tolerance to drought and/or salt stresses, respectively [31,32,33,34,35,36,37]. Previous study has shown that PtrWRKY51 responds to salt stress [38], but functional characterization of PtrWRKY51 in salt stress response remains unclear.
In this study, PtrWRKY51 was cloned from the Populus trichocarpa to investigate its function in salt stress in Arabidopsis. We analyzed its expression pattern in root, stem and leaf tissues and in response to salt stress. Moreover, the functional characterization of PtrWRKY51 in salt response was studied in Arabidopsis. Our results demonstrate that overexpression of PtrWRKY51 enhanced salt tolerance in Arabidopsis.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The solid Lloyd and McCown’s Woody Plant Basal Salts (WPM) medium was used for cultivation of the Populus trichocarpa in vitro [39]. After plantlet regeneration, they were individually transplanted and grown in pots (20.5 × 17 × 14.5 cm; top diameter/height/bottom diameter) containing a mixture of soil and vermiculite (2:1) and incubated in the greenhouse at 22 °C under a 16 h/8 h (light/dark) photoperiod (150 μmol m−2 s−1) and 70% relative humidity.
Arabidopsis Col-0 was selected as the wild-type control, and the mutant wrky51 (stock name SALK_022198.56.00.x) were obtained from Arabidopsis Biological Resource Center (ABRC; https://abrc.osu.edu/). Arabidopsis seeds were washed two times with water, then with 75% alcohol for 1 min followed by 1% NaClO within 10 min and five washes in distilled water. Seeds were sown on 1/2MS medium plates containing 30 g/L sucrose and 6 g/L agar. The seeded plates were vernalized at 4 °C in the dark for 2 days and then moved to 22 °C under 16/8 light/dark cycle. When Arabidopsis sprouted, it was transplanted at a density of four plants per pot (7 × 7 × 6.5 cm; top diameter/height/bottom diameter) with a mixture of soil and vermiculite (2:1) at 22 °C under 16/8 light/dark cycle and 70% relative humidity.
2.2. PtrWRKY51 Cloning and Transformation
Total RNA was extracted from the young leaves of Populus trichocarpa by using the RN38 EASYspin Plus Plant RNA Kit (Aidlab Biotech, Beijing, China). The first strand cDNA synthesis was reverse-transcribed using the Tiangen FastQuant RT Kit (Tiangen) according to the manufacturer’s instructions. The resultant cDNA was used as a template, and the PtrWRKY51 (Accession number: Potri.005G085200) sequence was amplified by PCR with gene-specific primers (Supplementary Table S1). The gene-specific primers were designed by using Primer Premier 5 software.
For overexpression of PtrWRKY51 and complementation of the Arabidopsis wrky51 mutant with PtrWRKY51, the PtrWRKY51 cDNA was cloned into the pCAMBIA-1301 binary vector under the control of the Cauliflower mosaic virus (CaMV) 35S promoter, and the plasmids were introduced into Agrobacterium tumefaciens strain GV3101 by heat shock. Then, the 35S: PtrWRKY51: GFP plasmid transformed into the Arabidopsis Col-0 and wrky51 mutant lines respectively by the floral dip method [40]. The transgenic lines were identified using half-strength MS plates containing 100 mg l−1 hygromycin. The third (T3) generation of seeds were used in the experiments to make sure hereditary stability in transgenic lines.
2.3. Molecular Verification of Transgenic Plants
Genomic DNA of Col-0 and transgenic Arabidopsis lines was extracted from leaves by using the CTAB method [41]. Transformants were identified by PCR amplification using the gene-specific primers PtrWRKY51-F and GFP-R (Supplementary Table S1). The transcript levels of PtrWRKY51 were quantified by Quantitative real-time PCR (RT-qPCR) in transgenic lines.
2.4. Quantitative Real-Time PCR Analysis
The ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems, Inc., Carlsbad, CA, USA) is used to perform Quantitative real-time PCR (RT-qPCR) experiments according to the manufacturer’s instructions. The 2−∆∆CT method [42] was used to calculate the relative expression level of PtrWRKY51, with poplar Actin employed as the internal control (Supplementary Table S1).
2.5. Physiological Experiments
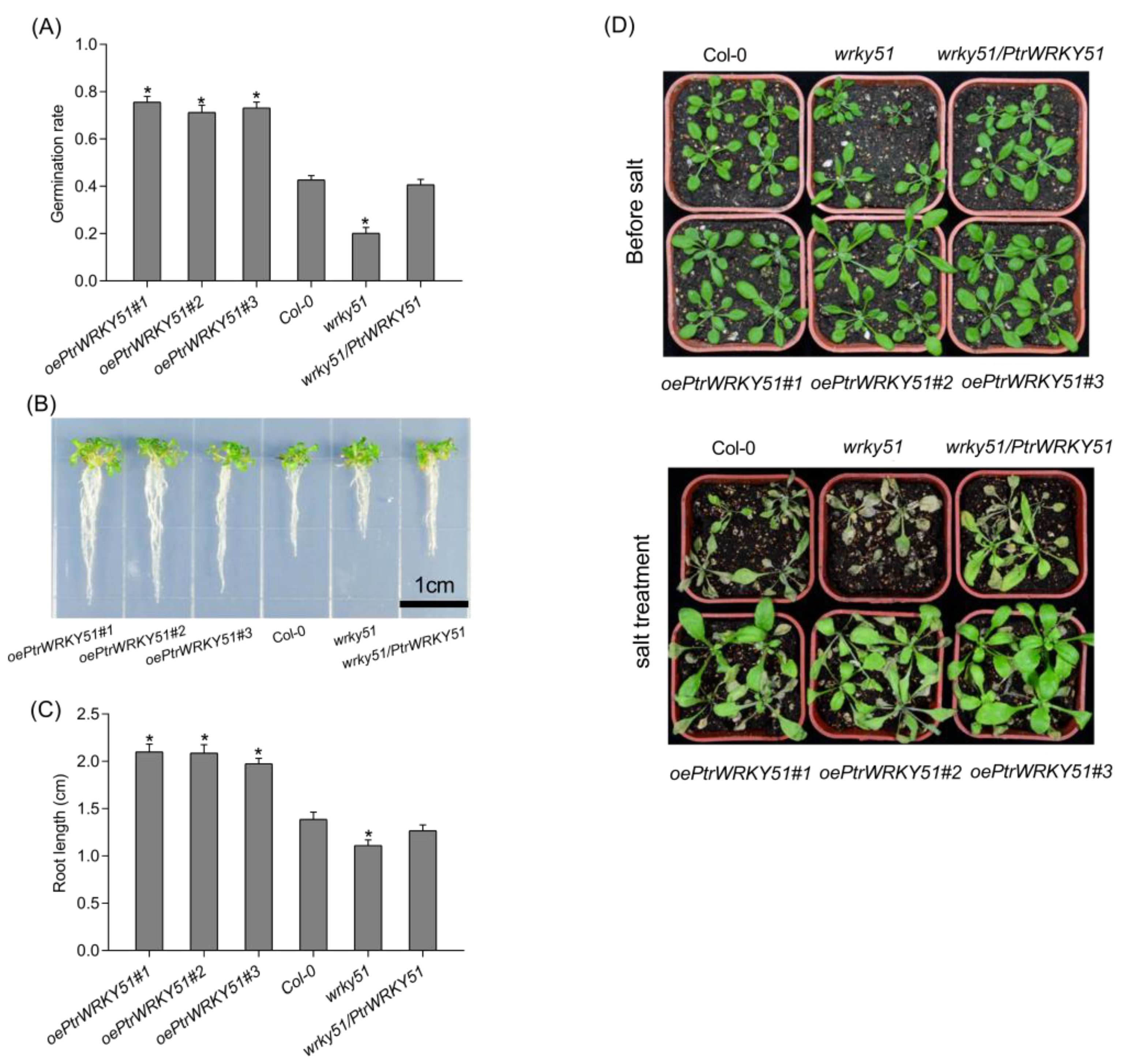
To determine germination rates of overexpressed PtrWRKY51(oePtrWRKY51), Col-0, wrky51 and wrky51/ PtrWRKY51 seeds, 100 seeds of different genotypes were seeded separately on the same 1/2MS medium with or without 200 mM NaCl stress and germination rates were recorded daily. When the seeds germinated, 10 plants of each genotype were selected and placed vertically on the medium with or without 200 mM NaCl stress to grow for 10 days, and the root length was recorded.
Net photosynthetic rate and transpiration rate were measured of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 plants by using an infrared gas analysis system (LI-COR 6400, Lincoln, NE, USA) as previously described [43,44]. Instantaneous leaf WUE was defined as the ratio of the rate of photosynthetic rate/transpiration rate [45].
Leaves of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 plants grown for 4 weeks were used for chlorophyll content determination with 80% acetone. A UV/visible spectrophotometer (YHB-061; GE Healthcare, Little Chalfont, Buckinghamshire, UK) was used to measure absorbance at 663 for chlorophyll a and 645 nm for chlorophyll b, and then the chlorophyll contents were calculated according to Lichtenthaler’s method [46].
2.6. Salt Experiments
For the salt treatments of Populus trichocarpa, 4-week-old seedlings were removed from the soil carefully and then placed in a salt solution containing 200 mM NaCl for 24 h. Subsequently, leaves were harvested at 0 h, 1 h, 3 h, 6 h, 8 h, 12 h and 24 h for RT-qPCR analysis.
Arabidopsis seeds of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 were seeded on 1/2MS medium and transplanted at a density of four plants per pot (7 × 7 × 6.5 cm; top diameter/height/bottom diameter) with a mixture of soil and vermiculite (2:1) at 22 °C under 16/8 light/dark cycle and 70% relative humidity after two true leaves had grown. It was cultured in the greenhouse with normal watered for two weeks, and then it was watered with 200 mM NaCl solution every 3 days for 15 days to observe its phenotype. The photosynthetic rates and instantaneous leaf WUE were measured at each stage as previously.
3. Results
3.1. The Analysis of Expression Pattern in PtrWRKY51
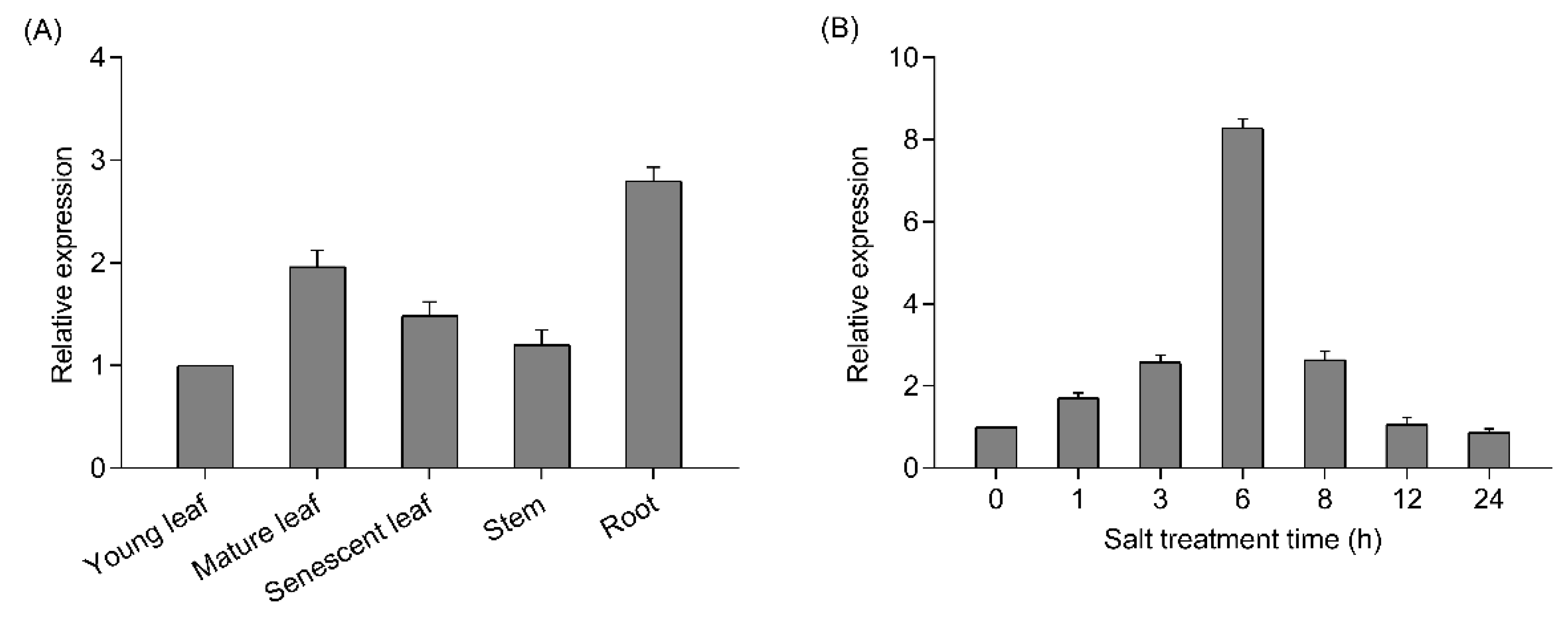
The tissue-specific expression of PtrWRKY51 transcripts in young leaf, mature leaf, senescent leaf, stem and root of P. trichocarpa was detected by RT-qPCR. The result showed that PtrWRKY51 transcript levels in mature leaves and root was higher than that in young leaf, senescent leaf and stem (Figure 1A). To study the response of PtrWRKY51 to salt stress, P. trichocarpa plants were subjected to salt stress and the PtrWRKY51 transcript levels were quantified by RT-qPCR. The results showed that the PtrWRKY51 transcript level gradually increased and peaked at 6 h of the NaCl treatments, and thereafter decreased (Figure 1B). These results indicated that PtrWRKY51 was mainly expressed in mature leaves and roots and up-regulated in response to salt stress.
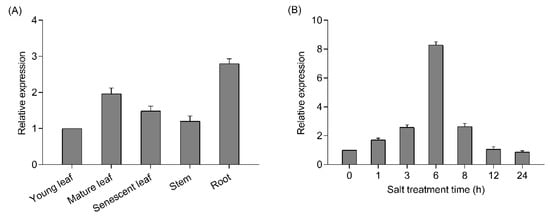
Figure 1.
Expression patterns of PtrWRKY51. (A)Transcript level of PtrWRKY51 in various organs including young leaf, mature leaf, senescent leaf, stem and root of P. trichocarpa. (B) RT-qPCR analysis of PtrWRKY51 transcript levels under salt conditions. Data are shown as mean ± SE (n = 6).
3.2. Identification of PtrWRKY51-Overexpressing Transgenic Plant
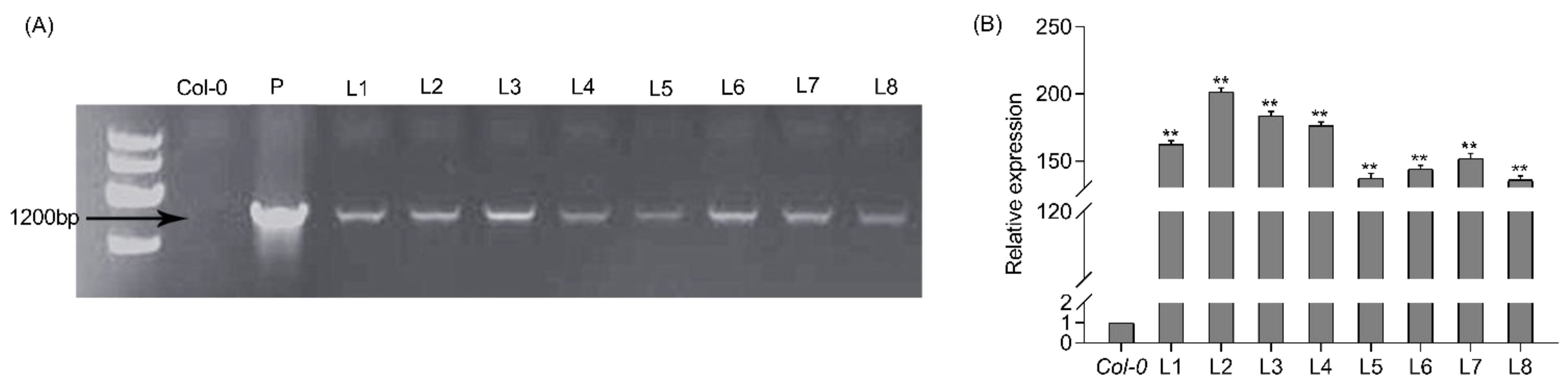
To further explore the function of PtrWRKY51 in salt stress conditions, the 35S: PtrWRKY51: GFP is transformed into Col-0 Arabidopsis. Eight transgenic lines (oePtrWRKY51 #1-7) were obtained and confirmed by PCR analyses using the combination of the gene-specific primers PtrWRKY51-F and GFP-R. The expected band was amplified in all transgenic plants but not in Col-0 plants (Figure 2A). In addition, RT-qPCR analysis of the above transgenic plants showed that PtrWRKY51 was overexpressed in all selected transgenic lines (Figure 2B). Then three of PtrWRKY51-overexpressing lines (oePtrWRKY51#1, #2, and #3) with higher expression level than other lines were selected for further analysis in salt tolerance.
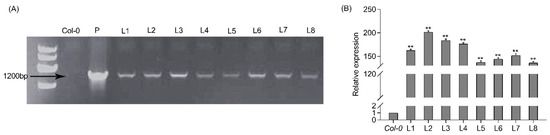
Figure 2.
Analysis of overexpressing PtrWRKY51 in transgenic plants. (A) Eight putatively transgenic plants of PtrWRKY51 were identified by PCR. (B) RT-qPCR was used to detect the transcription level of PtrWRKY51 in different lines. The data are represented as means ± SE (n = 6). The asterisks (**) represent a significant differences (** p < 0.01) compared to the control.
3.3. Plant Phenotype Analysis under Well-Watered Conditions
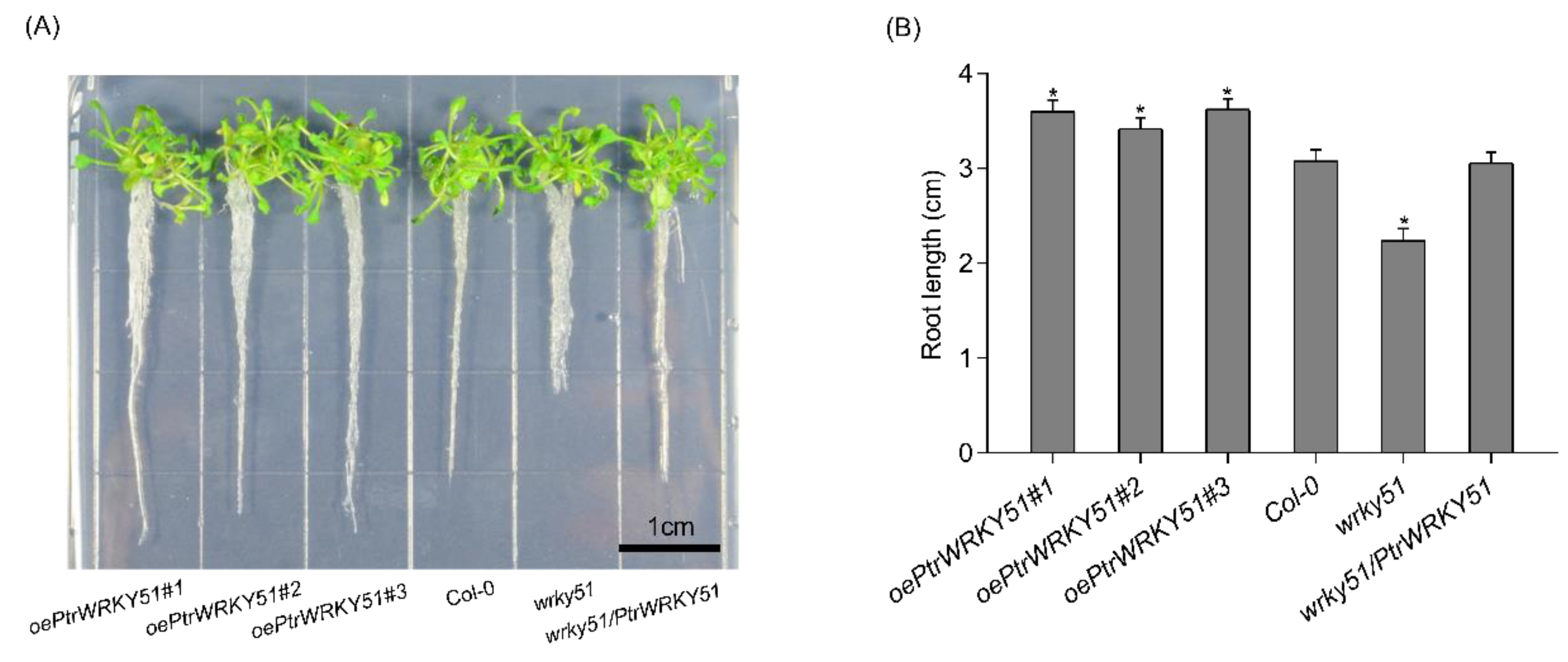
To explore the function of PtrWRKY51, the growth phenotype of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 plants were observed under normal conditions. The result showed that the primary root length of overexpressed plants was significantly longer than that of Col-0 (1.17-fold) and wrky51 (1.61-fold) after 10 days of vertical growth on 1/2 MS medium (Figure 3A,B). These results indicated that overexpression of PtrWRKY51 could promote root growth.
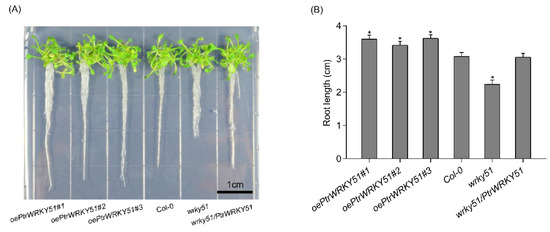
Figure 3.
Root length analysis of overexpressing PtrWRKY51 in plants under normal conditions. (A) Phenotypic analysis of the primary root length for 10 days on 1/2MS medium. Bars, 1 cm. (B) Difference in the primary root length after 10 days of vertical growth on 1/2MS medium. The data are represented as means ± SE (n = 12). The asterisks (*) represent significant differences (* p < 0.05) compared to the control.
3.4. Overexpression of PtrWRKY51 Enhanced WUE in Arabidopsis
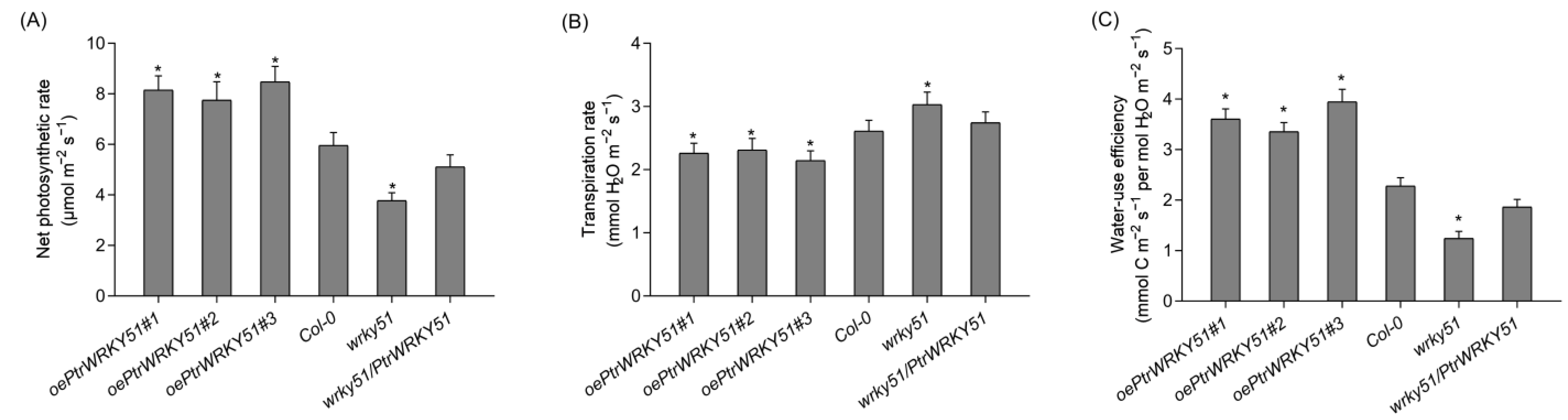
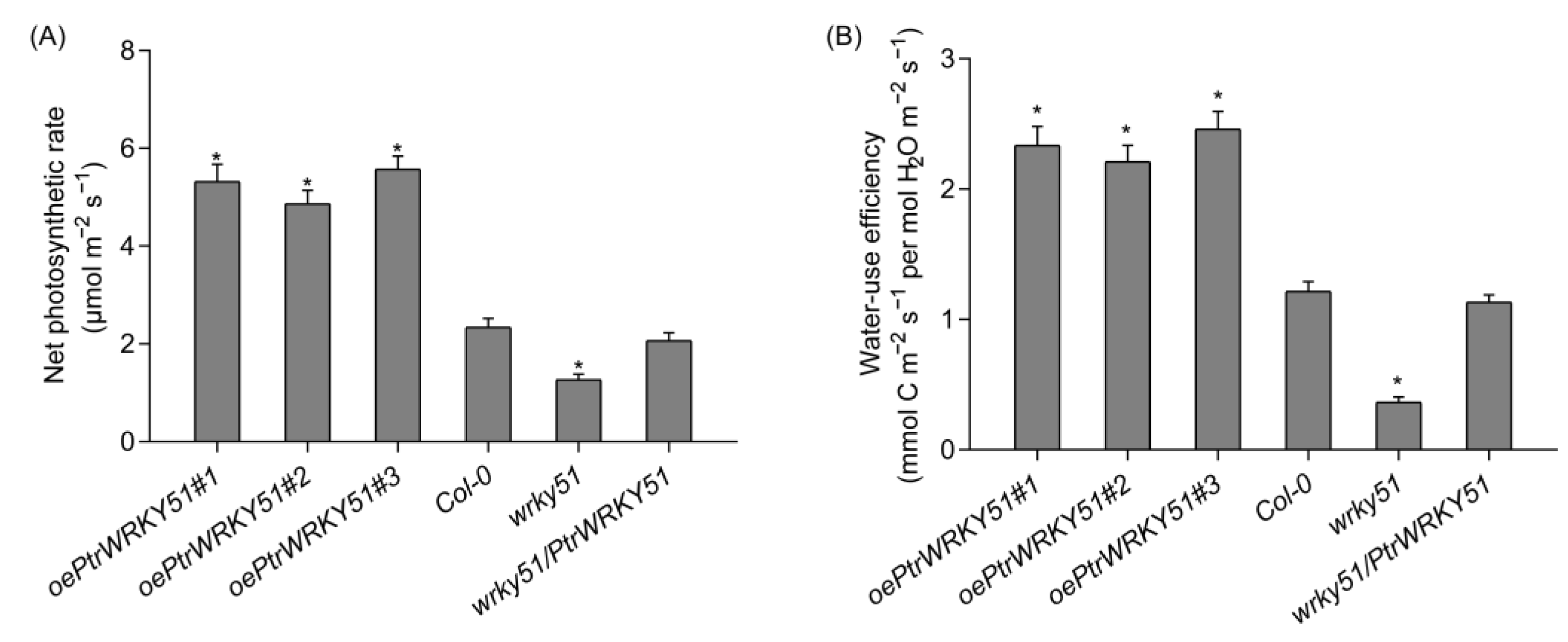
Photosynthetic physiological analysis that the net photosynthetic rates of oePtrWRKY51 lines was higher than that of Col-0 and wrky51 under normal conditions, and the net photosynthetic rate of wrky51/ PtrWRKY51 lines was equal to that of Col-0 (Figure 4A). In addition, the transpiration rate of each genotype was found that the oePtrWRKY51 lines showed slower rates than the Col-0 and wrky51 mutant (Figure 4B). As a result, based on the higher photosynthetic capability and lower transpiration level, the instantaneous WUE values of the oePtrWRKY51 lines were observably higher than that of Col-0 and wrky51 mutant (Figure 4C). In general, overexpression of PtrWRKY51 enhanced the WUE in Arabidopsis.
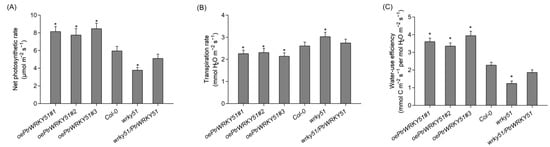
Figure 4.
Gas exchange analysis of overexpressing PtrWRKY51 lines showing higher instantaneous WUE in Arabidopsis. The Net photosynthesis rate (A), transpiration rate (B), instantaneous leaf WUE (C) of 3-week-old seedlings. The data are represented as means ± SE (n = 12). The asterisks (*) represent significant differences (* p < 0.05) compared to the control.
3.5. Overexpression of PtrWRKY51 Increases Salt Tolerance under Salt Stress Conditions
A series of experiments were conducted to explore the differences in salt tolerance of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 plants. The seeds of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 were sown on 1/2 MS culture containing 200 mM NaCl to observe germination rates. After 4 days, the oePtrWRKY51 plants had a higher germination rate (73.4%) than the Col-0 (42.8%) and mutant (20.2%) plants (Figure 5A). In addition, the primary root length of the seedlings was also different after 10 days of vertical growth under salt stress conditions. The primary root length of oePtrWRKY51 plants was significantly longer (1.48- and 1.85- fold, respectively) than that of Col-0 and mutant (Figure 5B,C). After the seedlings were transplanted to soil, the salt stress was imposed for 15 days. As expected, the Col-0 and wrky51 mutant plants showed more severe wilting than over-expressing plants, especially the wrky51 mutants, whereas those of the oePtrWRKY51 lines continued to show development and growth (Figure 5D).
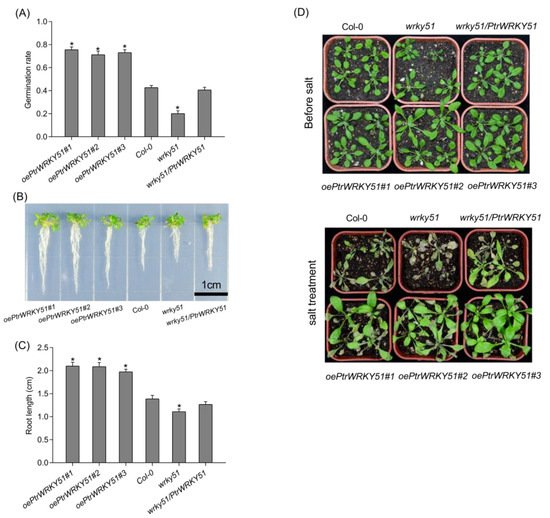
Figure 5.
Overexpression of PtWRKY51 confers salt tolerance in Arabidopsis. (A) Difference in germination ratio among oePtWRKY51, Col-0, wrky51, and wrky51/ PtWRKY51 plants grown on 1/2 MS medium with 200 mM NaCl. (B) Morphological comparisons of the primary root length for 10 d under 1/2 MS medium with 200 mM NaCl conditions. (C) Difference in the primary root length after 10 days of vertical growth on 1/2 MS medium with 200 mM NaCl conditions. (D) Morphological differences in salt treatments experiments. The data are represented as means ± SE (n = 12). The asterisks (*) represent significant differences (* p < 0.05) compared to the control.
The net photosynthesis rate and WUE of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 plants were measured and calculated under the salt treatments conditions (Figure 6A,B). The result showed that compared with the Col-0 and wrky51 mutant plants, the oePtrWRKY51 lines could maintain a higher photosynthetic rate and have a higher instantaneous leaf WUE under salt stress conditions.
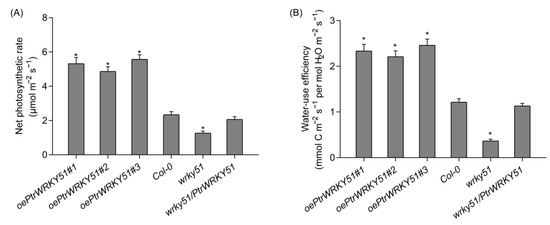
Figure 6.
Physiological analysis of over-expressing PtWRKY51 lines under salt stress conditions. Difference in net photosynthetic rate (A) and instantaneous leaf WUE (B) among oePtWRKY51, Col-0, wrky51, and wrky51/ PtWRKY51 plants under salt stress conditions. The data are represented as means ± SE (n = 12). The asterisks (*) represent significant differences (* p < 0.05) compared to the control.
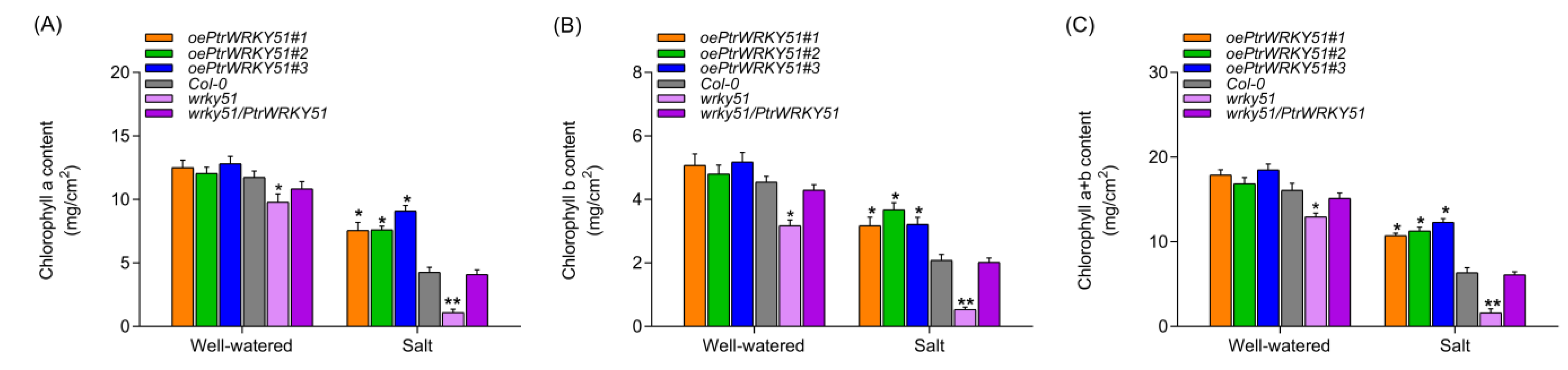
To further assess the potential biological functions of PtrWRKY51 in regulation of salt tolerance, the chlorophyll a, chlorophyll b, and total chlorophyll contents of oePtrWRKY51, Col-0, wrky51 and wrky51/ PtrWRKY51 plants were also measured under normal and salt stress conditions. The result showed that there was no significant difference in chlorophyll content between the oePtrWRKY51 and the Col-0 plants, but there was a difference between them and the wrky51 mutant under normal conditions. However, under salt stress conditions, the chlorophyll content of oePtrWRKY51 plants was markedly higher than that of Col-0 and wrky51 mutant (Figure 7), indicating that the oePtrWRKY51 plants showed a better ability to absorb light energy compared with Col-0 and wrky51 mutant, thus the oePtrWRKY51 plants maintained a higher photosynthetic rate under salt stress conditions. Therefore, the expression of PtrWRKY51 was beneficial for plant growth under salt stress conditions.
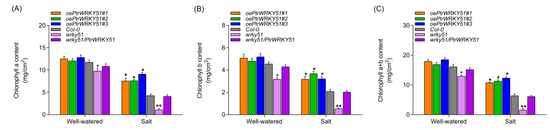
Figure 7.
The chlorophyll content analysis of over-expressing PtWRKY51 plants under salt treatment conditions. Determine of the content of chlorophyll a (A), chlorophyll b (B) and chlorophyll a + b (C) among oePtWRKY51, Col-0, wrky51, and wrky51/ PtWRKY51 plants under salt stress conditions. The data are represented as means ± SE (n = 12). The asterisks (* and **) represent significant differences (* p < 0.05 and ** p < 0.01) compared to the control.
4. Discussion
Salt stress has seriously affected and inhibited plant growth and development. Stress-induced transcription factors (TFs) are important pivotal regulators in response to stress. They can bind membrane protein acting elements and promote related genes expression in response to stress to reduce damage of the stress. WRKY is a plant-specific zinc finger type transcription factor. It is involved in a wide variety of biotic and abiotic stress responses [20,38,47,48,49,50,51]. Studies have shown that OsWRKY51 and OsWRKY71 regulate GA and ABA signal transduction in seed germination [52]. In addition, AtWRKY51 was involved in JA-induced defense response [53]. Nevertheless, little was known about the function of WRKY51 in abiotic, especially in salt stress. Previous studies have shown that PtrWRKY51 responds to salt [38]. However, the function of PtrWRKY51 in salt stress remains unclear. In the study, PtrWRKY51 was cloned from poplar and genetically modified in Arabidopsis to determine its potential role resistance to salt stress.
To explore the role of genes in response to biological or abiotic stress, tissue-specific gene expression patterns play a crucial role in evaluating the potential function of genes [54,55,56]. Therefore, a novel idea was provided to investigate the potential functions of PtrWRKY51 by studying the tissue-specific expression. Analysis of transcription patterns showed that PtrWRKY51 was predominantly expressed in mature leaves and root (Figure 1A). In addition, the result showed that the primary root length of oePtrWRKY51 plants was significantly longer than that of Col-0 and wrky51 on 1/2 MS medium (Figure 3A,B). Moreover, PtrWRKY51 were up-regulated by salt stress (Figure 1B), and oePtrWRKY51 plants have a prominently longer root that of Col-0 and wrky51 on 1/2 MS medium with 200 mM NaCl (Figure 5B,C). These findings suggested that PtrWRKY51 plays a key role in the regulation of growth and salt tolerance in plants.
In generally, the growth status and external morphology of a plant can reflect the stress degree in the plant. Seed germination is a critically and extremely sensitive period in plant growth, and most plants in salt environment have a negative impact on their growth rate. In the germination stage, dry seed germination is promoted through water expansion, soil water potential is decreased in salt soil, and the absorption of water by seeds is inhibited, which seriously affects the germination rate, vigor index and germination index of seeds, which leading to the reduction of seed emergence rate [8,9]. The result showed that the oePtrWRKY51 plants had a higher germination rate than the Col-0 and mutant plants under salt treatments (Figure 5A). Therefore, overexpression of PtrWRKY51 is beneficial to plant in growth under salt stress conditions. Salt stress not only inhibited seed germination, but also significantly inhibited the growth of seedlings. Under salt stress conditions, the root structure of the plant was significantly damaged, and the root length was dramatically reduced, which affected the transport of water and nutrients from the root system to the aboveground part, which led to the obstruction of organic synthesis, and ultimately affected the growth and development of plants [57]. In this study, the primary root length of oePtrWRKY51 lines was significantly longer than that of Col-0 and wrky51 mutant under salt stress conditions, which was conducive to plant growth and development (Figure 5B, C). In summary, these results further supported the conclusion that overexpression of PtrWRKY51 enhances salt tolerance in salt stress conditions.
Photosynthesis is one of the most sensitive physiological processes in plant response to stress [58]. Under normal growth conditions, the photosynthesizing organic matter of plants is enough to supply their needs, but under some special stress, the photosynthesis of plants will be affected, and they cannot synthesize enough organic matter to ensure the growth and breeding in plants. These stresses include cold, drought, high temperature, salt and so on. Among them, the damage area of salt stress is wider, and the damage degree of salt stress is deeper. The net photosynthetic rate analysis showed that photosynthesis was higher in the oePtrWRKY51 lines compared to Col-0 and wrky51 mutant under normal conditions, while the transpiration rate was lower than that of Col-0 and wrky51 (Figure 4A,B). Therefore, the instantaneous WUE of the oePtrWRKY51 lines were significantly higher than those of Col-0 and wrky51 mutant lines (Figure 4C). WUE has been considered as one of the key factors which affect plant growth, and high WUE can promote plant growth [36]. WUE is also an important physiological index to assess plant stress resistance [59,60]. Under salt stress, the net photosynthetic rate and WUE of plants decreased, but oePtrWRKY51 lines still had a higher net photosynthetic rate and WUE (Figure 6). Therefore, overexpression of PtrWRKY51 improve plant growth and salt tolerance.
The chlorophyll content of leaves is closely related to photosynthesis in plants [61]. Salt stress can damage the ultrastructure of chloroplasts and affect the normal function of cells, which affecting the growth and development of plants [62]. We observed that under normal growth conditions, there was no significant difference in chlorophyll content between oePtrWRKY51 and Col-0 plants, but both were higher than that of wrky51 mutant plants. Under salt stress, chlorophyll content of oePtrWRKY51 plants was significantly higher than that of Col-0 and wrky51 mutant, which suggested oePtrWRKY51 lines have a growth advantage under salt stress conditions. Taken together, these data indicate that PtrWRKY51 is a promising gene target in increasing the tolerance of plants under salt stress conditions.
5. Conclusions
In this study, we illustrate the functional characterization of the poplar WRKY transcription factor PtrWRKY51 in the salt response. A PtrWRKY51 was isolated from Populus trichocarpa. RT-qPCR analysis revealed that PtrWRKY51 was mainly expressed in mature leaves and root. In addition, PtrWRKY51 is induced by salt stress. Overexpression of PtrWRKY51 in Arabidopsis improved salt tolerance. Consistently, overexpression of PtrWRKY51 exhibit an increase in seed germination rate, root length, photosynthetic rate, instantaneous leaf WUE, chlorophyll content under salt stress conditions. Taken together, our data indicate that PtrWRKY51 is a potential candidate gene in the improvement of salt tolerance in poplar by biotechnological strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f14020191/s1, Table S1: Primer sequences used for cloning of PtrWRKY51 cDNA and RT-PCR.
Author Contributions
Conceptualization, Y.Z. (Yangyan Zhou), Q.L. and Y.Z. (Yue Zhang); Formal analysis, Y.Z. (Yangyan Zhou) and Q.L.; Funding acquisition, Y.Z. (Yangyan Zhou); Investigation, Y.Z. (Yangyan Zhou) and Q.L.; Methodology, Y.Z. (Yangyan Zhou); Supervision, Y.Z. (Yangyan Zhou), Q.L. and Y.Z. (Yue Zhang); Writing—original draft, Y.Z. (Yangyan Zhou); Writing—review & editing, Y.Z. (Yue Zhang). All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by China Postdoctoral Science Foundation (2022M710215) and the Natural Science Foundation of Shandong Province (ZR2022QC162).
Data Availability Statement
The data set used in this study can be made available from the authors upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yang, Y.Q.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Fasoli, M.; Tornielli, G.B.; Dal Santo, S.; Pezzotti, M.; Zhang, L.S.; Cai, B.; Cheng, Z.M. The evolutionary history and diverse physiological roles of the grapevine calcium-dependent protein kinase gene family. PLoS ONE 2013, 8, e80818. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Beltran, J.; Manzur, C.L. An overview of the word’s salinity issues and FAO’s strategy to address this issue. In Proceedings of the International Salinity Forum Proceedings, Riverside, CA, USA, 25–27 April 2005; Volume 5, pp. 311–313. [Google Scholar]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Ouhibi, C.; Attia, H.; Rebah, F.; Msilini, N.; Chebbi, M.; Aarrouf, J.; Urban, L.; Lachaal, M. Salt stress mitigation by seed priming with UV-C in lettuce plants: Growth, antioxidant activity and phenolic compounds. Plant Physiol. Biochem. 2014, 83, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Bo, C.; Chen, H.W.; Luo, G.W.; Li, W.; Zhang, X.G.; Ma, Q.; Cheng, B.J.; Cai, R.H. Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice. Plant Cell Rep. 2020, 39, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Keyster, M.; Klein, A.; Ludidi, N. Caspase-like enzymatic activity and the ascorbate-glutathione cycle participate in salt stress tolerance of maize conferred by exogenously applied nitric oxide. Plant Signal Behav. 2012, 7, 349–360. [Google Scholar] [CrossRef]
- Hu, H.; Liu, H.; Liu, F. Seed germination of hemp (Cannabis sativa L.) cultivars responds differently to the stress of salt type and concentration. Ind. Crops Prod. 2018, 123, 254–261. [Google Scholar] [CrossRef]
- Wang, W.B.; Kim, Y.H.; Lee, H.S.; Kim, K.Y.; Deng, X.P.; Kwak, S.S. Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 2009, 47, 570–577. [Google Scholar] [CrossRef] [PubMed]
- İbrahimova, U.; Kumari, P.; Yadav, S.; Rastogi, A.; Antala, M.; Suleymanova, Z.; Zivcak, M.; Tahjib-Ul-Arif, M.; Hussain, S.; Abdelhamid, M.; et al. Progress in understanding salt stress response in plants using biotechnological tools. J. Biotechnol. 2021, 329, 180–191. [Google Scholar] [CrossRef]
- Chen, Y.; Hoehenwarter, W. Changes in the Phosphoproteome and Metabolome Link Early Signaling Events to Rearrangement of Photosynthesis and Central Metabolism in Salinity and Oxidative Stress Response in Arabidopsis. Plant Physiol. 2015, 169, 3021–3033. [Google Scholar] [CrossRef]
- López-Climent, M.F.; Arbona, V.; Pérez-Clemente, R.M.; Gómez-Cadenas, A. Relationship between salt tolerance and photosynthetic machinery performance in citrus. Environ. Exp. Bot. 2008, 62, 176–184. [Google Scholar] [CrossRef]
- Mittal, S.; Kumari, N.; Sharma, V. Differential response of salt stress on Brassica juncea: Photosynthetic performance, pigment, proline, D1 and antioxidant enzymes. Plant Physiol. Biochem. 2012, 54, 17–26. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wu, Y.H.; Zhao, Q.; Wang, B.; Liu, Q.L.; Zhang, L. Chrysanthemum DgWRKY2 Gene Enhances Tolerance to Salt Stress in Transgenic Chrysanthemum. Int. J. Mol. Sci. 2018, 19, 2062. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP Research Group. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Marè, C.; Mazzucotelli, E.; Crosatti, C.; Francia, E.; Stanca, A.M.; Cattivelli, L. Hv-WRKY38: A new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 2004, 55, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA binding proteins bind to a conserved cis-element in the promoters of a-Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef] [PubMed]
- de Pater, S.; Greco, V.; Pham, K.; Memelink, J.; Kijne, J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996, 24, 4624–4631. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.P.; Jing, S.J.; Fu, J.; Li, L.; Yu, D.Q. Cloning and analysis of expression profile of 13 WRKY genes in rice. Chin. Sci. Bull. 2004, 49, 2159–2168. [Google Scholar] [CrossRef]
- Wu, H.L.; Ni, Z.F.; Yao, Y.Y.; Guo, G.G.; Sun, Q.X. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat (Triticum aestivem L.). Prog. Nat. Sci. 2008, 18, 697–705. [Google Scholar] [CrossRef]
- Qiu, Y.P.; Yu, D.Q. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2008, 65, 35–47. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Yu, D.Q.; Li, G.X.; Zhang, S.Q.; Zheng, S.J. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014, 79, 13–27. [Google Scholar] [CrossRef]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef]
- Ma, J.; Gao, X.; Liu, Q.; Shao, Y.; Zhang, D.; Jiang, L.; Li, C. Overexpression of TaWRKY146 Increases Drought Tolerance through Inducing Stomatal Closure in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 2036. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, H.; Han, G.; Cai, R.; Pan, F.; Xiang, Y. A moso bamboo WRKY gene PeWRKY83 confers salinity tolerance in transgenic Arabidopsis plants. Sci. Rep. 2017, 7, 11721. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Yang, J.F.; Li, M.; Xu, Z.S.; Fu, J.D. The Maize WRKY Transcription Factor ZmWRKY40 Confers Drought Resistance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cheng, J.; Sun, X.; Zhao, T.; Li, M.; Wang, Q.; Li, S.; Xin, H. Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep. 2018, 37, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.Z.; Duan, Y.J.; Yin, J.; Ye, S.L.; Zhu, J.R.; Zhang, F.Q.; Lu, W.X.; Fan, D.; Luo, K.M. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 65, 6629–6644. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lu, S.; Chen, Z.Z.; Lourenco, R.; Chiang, V.L. Genetic transformation of Populus trichocarpa genotype Nisqually-1: A functional genomic tool for woody plants. Plant Cell Physiol. 2006, 47, 1582–1589. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of ctab dna extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.; Dong, Y.; Zhao, Y.; Geng, A.; Xia, X.; Yin, W. PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 2016, 14, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tang, S.; An, Y.; Zheng, D.; Xia, X.; Yin, W. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 2013, 64, 4589–4601. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.C.; Cowan, I.R.; Farquhar, G.D. Leaf Conductance in Relation to Assimilation in Eucalyptus pauciflora Sieb. ex Spreng: Influence of Irradiance and Partial Pressure of Carbon Dioxide. Plant Physiol. 1978, 62, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids-pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Parinita, A.; Reddy, M.P.; Jitendra, C. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Et Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.S.; Liu, C.T.; Zhang, Y.; Meng, X.P.; Zhou, X.; Chu, C.C.; Wang, X.P. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef]
- Zhu, H.; Zhou, Y.Y.; Zhai, H.; He, S.Z.; Zhao, N.; Liu, Q.C. A Novel Sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis. Biomolecules 2020, 10, 506. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Yang, G.; Komatsu, S.; Shen, Q.J. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006, 46, 231–242. [Google Scholar] [CrossRef]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.; Tayrose, G.; Holt, B.F. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef]
- Horváth, B.M.; Magyar, Z.; Zhang, Y.; Hamburger, A.W.; Bakó, L.; Visser, R.G.; Bachem, C.W.; Bögre, L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006, 25, 4909–4920. [Google Scholar] [CrossRef]
- Sun, W.; Xu, X.; Zhu, H.; Liu, A.; Liu, L.; Li, J.; Hua, X. Comparative transcriptomic profiling of a salt-tolerant wild tomato species and a salt-sensitive tomato cultivar. Plant Cell Physiol. 2010, 51, 997–1006. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.; Rubio, F.; Garcia-Sanchez, F.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef]
- Karaba, A.; Dixit, S.; Greco, R.; Aharoni, A.; Trijatmiko, K.R.; Martinez, N.M.; Krishnan, A.; Nataraja, K.N.; Udayakumar, M.; Pereira, A. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. USA 2007, 104, 15270–15275. [Google Scholar] [CrossRef]
- An, Y.; Han, X.; Tang, S.; Xia, X.; Yin, W. Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. 2014, 119, 313–327. [Google Scholar] [CrossRef]
- Peharec Štefanić, P.; Koffler, T.; Adler, G.; Bar-Zvi, D. Chloroplasts of salt-grown Arabidopsis seedlings are impaired in structure, genome copy number and transcript levels. PLoS ONE 2013, 8, e82548. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).