Genetic Monitoring of Alnus glutinosa Natural Populations Using Two Generation Cohorts
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. DNA and Selected Marker Quality
3.2. Genetic Diversity of Different Age Cohorts in A. glutinosa Populations
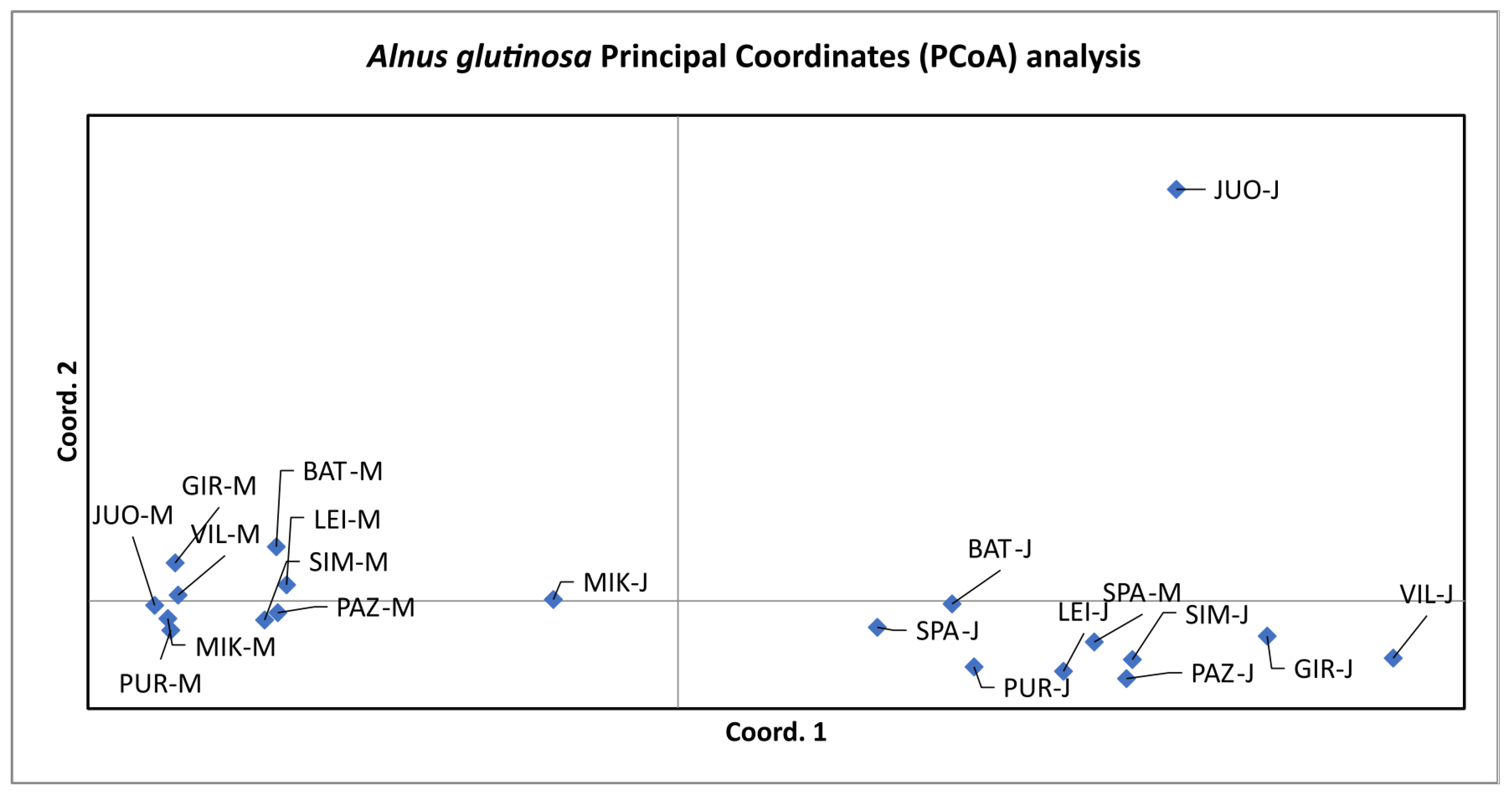
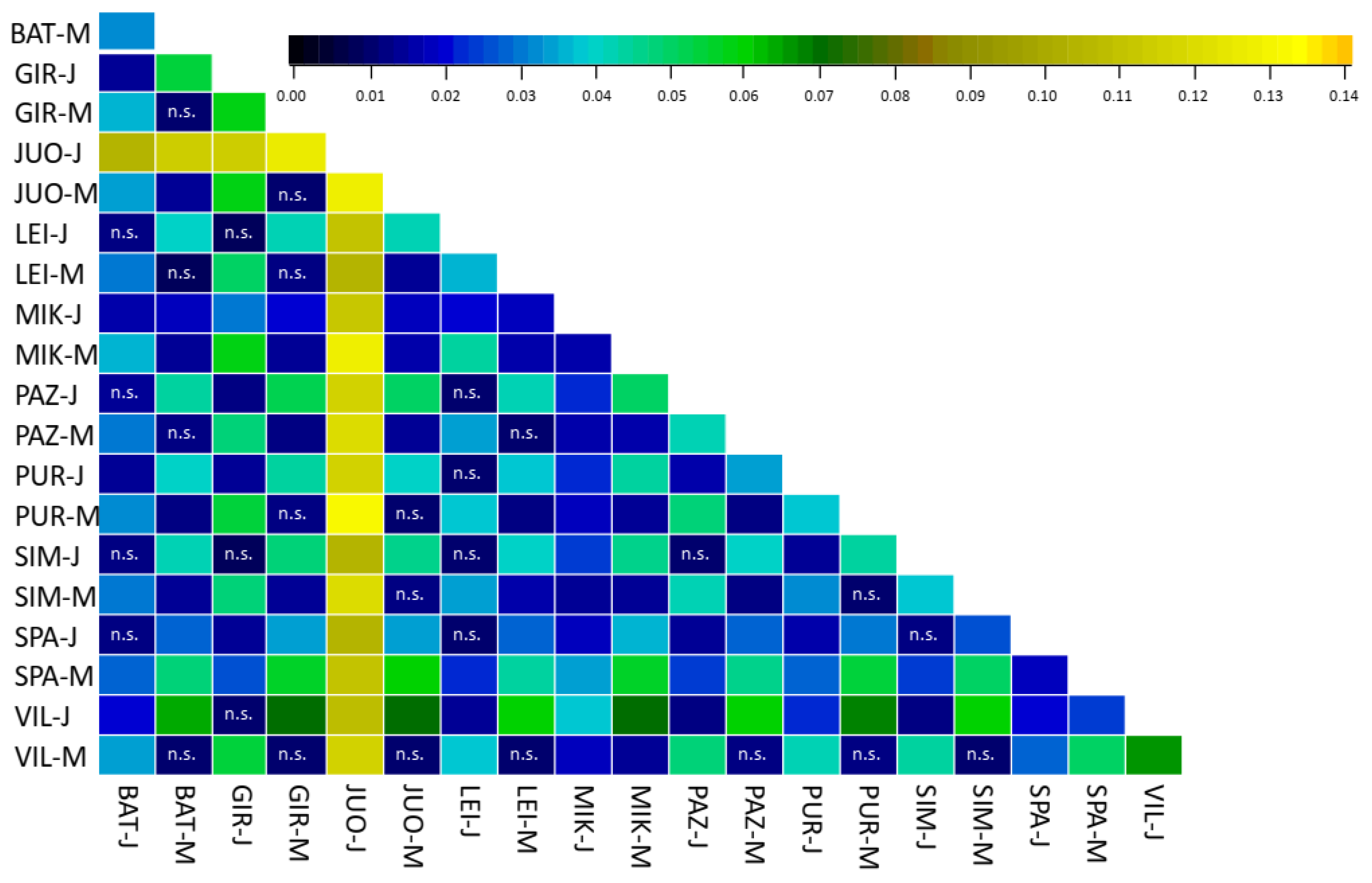
3.3. Population Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kremer, A.; Potts, B.M.; Delzon, S. Genetic divergence in forest trees: Understanding the consequences of climate change. Funct. Ecol. 2014, 28, 22–36. [Google Scholar] [CrossRef]
- Barry, W.; Brook, B.W.; Sodhi, N.S.; Bradshaw, C.J.A. Synergies among extinction drivers under global change. Trends Ecol. Evol. 2008, 23, 453–460. [Google Scholar] [CrossRef]
- Hof, C.; Levinsky, I.; Araújo, M.B.; Rahbek, C. Rethinking species’ ability to cope with rapid climate change. Glob. Chang. Biol. 2011, 17, 2987–2990. [Google Scholar] [CrossRef]
- Meier, E.S.; Lischke, H.; Schmatz, D.R.; Zimmermann, N. Climate, competition and connectivity affect future migration and ranges of European trees. Glob. Ecol. Biogeogr. 2012, 21, 164–178. [Google Scholar] [CrossRef]
- Vranckx, G.; Jacquemyn, H.; Muys, B.; Honnay, O. Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv. Biol. 2012, 26, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Aravanopoulos, F.A.; Bajc, M.; Fussi, B.; Kraigher, H. Introduction. In Manual for Forest Genetic Monitoring; Bajc, M., Aravanopoulos, F., Westergren, M., Fussi, B., Kavaliauskas, D., Alizoti, P., Kiourtsis, F., Kraigher, H., Eds.; Slovenian Forestry Institute, Silva Slovenica Publishing Centre: Ljubljana, Slovenia, 2020; pp. 11–14. [Google Scholar]
- Aravanopoulos, F.A. Genetic monitoring in natural perennial plant populations. Botany, 2011, 89, 75–81. [Google Scholar] [CrossRef]
- McVean, D.N. Alnus glutinosa (L.) Gaertn. J. Ecol. 1953, 41, 4472466. [Google Scholar] [CrossRef]
- Chambers, F.M.; Elliott, L. Spread and Expansion of Alnus Mill. In the British Isles: Timing, Agencies and Possible Vectors. J. Biogeogr. 1989, 16, 541–550. [Google Scholar] [CrossRef]
- Kajba, D.; Gračan, J. EUFORGEN Technical Guidelines for Genetic Conservation and Use for Black Alder (Alnus glutinosa); International Plant Genetic Resources Institute: Rome, Italy, 2003; pp. 1–4. [Google Scholar]
- Glenz, C.; Schlaepfer, R.; Iorgulescu, I.; Kienast, F. Flooding tolerance of Central European tree and shrub species. For. Ecol. Manag. 2006, 235, 1–13. [Google Scholar] [CrossRef]
- Houston Durrant, T.; de Rigo, D.; Caudullo, G. Alnus glutinosa in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Public Office EU: Luxembourg, 2016; p. e01f3c0+. [Google Scholar]
- Ozolinčius, R. Alnus gentis. Alnus glutinosa. [Genus Alnus. Alnus glutinosa]. In Lietuvos Dendroflora [Dendroflora of Lithuania]; Navasaitis, M., Ozolinčius, R., Smaliukas, D., Balevičienė, J., Eds.; Lututė: Vilnius, Lithuania, 2003; p. 576. [Google Scholar]
- Havrdová, A.; Douda, J.; Krak, K.; Vít, P.; Hadincová, V.; Zákravský, P.; Mandák, B. Higher genetic diversity in recolonized areas than in refugia of Alnus glutinosa triggered by continent-wide lineage admixture. Mol. Ecol. 2015, 24, 4759–4777. [Google Scholar] [CrossRef]
- Cox, K.; Vanden Broeck, A.; Van Calster, H.; Mergeay, J. Temperature-related natural selection in a wind-pollinated tree across regional and continental scales. Mol. Ecol. 2011, 20, 2724–2738. [Google Scholar] [CrossRef] [PubMed]
- Lepais, O.; Muller, S.D.; Ben Saad-Limam, S.; Benslama, M.; Rhazi, L.; Belouahem-Abed, D.; Daoud-Bouattour, A.; Gammar, A.M.; Ghrabi-Gammar, Z.; Bacles, C.F.E. High genetic diversity and distinctiveness of rear-edge climate relicts maintained by ancient tetraploidisation for Alnus glutinosa. PLoS ONE 2013, 8, 75029. [Google Scholar] [CrossRef] [PubMed]
- Mingeot, D.; Husson, C.; Mertens, P.; Watillon, B.; Bertin, P.; Druart, P. Genetic diversity and genetic structure of black alder(Alnus glutinosa [L.] Gaertn) in the Belgium-Luxembourg-Francecross-border area. Tree Genet. Genomes 2016, 12, 24. [Google Scholar] [CrossRef]
- Beatty, G.E.; Montgomery, W.I.; Tosh, D.G.; Provan, J. Genetic provenance and best practice woodland management: A case study in native alder (Alnus glutinosa). Tree Genet. Genomes 2015, 11, 1–7. [Google Scholar] [CrossRef]
- Mandák, B.; Vít, P.; Krak, K.; Trávníček, P.; Havrdová, A.; Hadincová, V.; Zákravský, P.; Jarolímová, V.; Bacles, C.F.; Douda, J. Flow cytometry, microsatellites and niche models reveal the origins and geographical structure of Alnus glutinosa populations in Europe. Ann. Bot. 2016, 117, 107–120. [Google Scholar] [CrossRef] [PubMed]
- De Kort, H.; Vandepitte, K.; Bruun, H.H.; Closset-Kopp, D.; Honnay, O.; Mergeay, J. Landscape genomics and a common garden trial reveal adaptive differentiation to temperature across Europe in the tree species Alnus glutinosa. Mol. Ecol. 2014, 23, 4709–4721. [Google Scholar] [CrossRef]
- Caudullo, G.; Welk, E.; San-Miguel-Ayanz, J. Chorological maps for the main European woody species. Data Brief 2017, 12, 662–666. [Google Scholar] [CrossRef]
- Forest resources. In Lithuanian Statistical Yearbook of Forestry; State Forestry Srvice: Kaunas, Lithuania, 2021; pp. 12–25.
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pedunculated oak investigated with an efficient PCR method. Ther. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef]
- Lepais, O.; Bacles, C.F.E. De novo discovery and multiplexed amplification of microsatellite markers for black alder (Alnus glutinosa) and related species using SSR-enriched shotgun pyrosequencing. J. Hered. 2011, 102, 627–632. [Google Scholar] [CrossRef]
- Drašnarová, A.; Krak, K.; Vít, P.; Doudová, J.; Douda, J.; Hadincová, V.; Zákravský, P.; Mandák, B. Cross-amplification and multiplexing of SSR markers for Alnus glutinosa and A. incana. Tree Genet. Genomes 2014, 10, 865–873. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchison, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Smouse, P.E.; Banks, S.C.; Peakall, R. Converting quadratic entropy to diversity: Both animals and alleles are diverse, but some are more diverse than others. PLoS ONE 2017, 12, e0185499. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary N e using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Dakin, E.E.; Avise, J.C. Microsatellite null alleles in parentage analysis. Heredity 2004, 93, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Lian, C.; Hogetsu, T. Development of microsatellite markers in white birch (Betula platyphylla var. japonica). Mol. Ecol. 2002, 3, 413–415. [Google Scholar] [CrossRef]
- Gürcan, K.; Mehlenbacher, S.A.; Botta, R.; Boccacci, P. Development, characterization, segregation, and mapping of microsatellite markers for European hazelnut (Corylus avellana L.) from enriched genomic libraries and usefulness in genetic diversity studies. Tree Genet. Genomes 2010, 6, 513–531. [Google Scholar] [CrossRef]
- Kulju, K.K.M.; Pekkinen, M.; Varvio, S. Twenty-three microsatellite primer pairs for Betula pendula (Betulaceae). Mol. Ecol. Notes 2004, 4, 471–473. [Google Scholar] [CrossRef]
- Tsuda, Y.; Ueno, S.; Ide, Y.; Tsumura, Y. Development of 14 ESTSSRs for Betula maximowicziana and their applicability to related species. Conserv. Genet. 2009, 10, 661–664. [Google Scholar] [CrossRef]
- Ogyu, K.; Tsuda, Y.; Sugaya, T.; Yoshimaru, H.; Ide, Y. Identification and characterization of microsatellite loci in Betula maximowicziana Regel. Mol. Ecol. Notes 2003, 3, 268–269. [Google Scholar] [CrossRef]
- Tsuda, Y.; Ueno, S.; Ranta, J.; Salminen, K.; Ide, Y.; Shinohara, K.; Tsumura, Y. Development of 11 EST-SSRs for Japanese white birch, Betula platyphylla var. japonica and their transferability to related species. Conserv. Genet. 2009, 10, 1385–1388. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.G.; Samis, K.E.; Lougheed, S.C. Genetic variation across species’ geographical ranges: The central-marginal hypothesis and beyond. Mol. Ecol. 2008, 17, 1170–1188. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Aguinagalde, I.; de Beaulieu, J.L.; Bittkau, C.; Brewer, S.; Cheddadi, R.; Ennos, R.; Fineschi, S.; Grivet, D.; Lascoux, M.; et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science 2003, 300, 1563–1565. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Brewer, S.; Bordács, S.; Burg, K.; Cheddadi, R.; Coart, E.; Cottrell, J.; Csaikl, U.M.; van Dam, B.; Deans, J.D.; et al. Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For. Ecol. Manag. 2002, 156, 49–74. [Google Scholar] [CrossRef]
- Heuertz, M.; Fineschi, S.; Anzidei, M.; Pastorelli, R.; Salvini, D.; Paule, L.; Frascaria-Lacoste, N.; Hardy, O.J.; Vekemans, X.; Vendramin, G.G. Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol. Ecol. 2004, 13, 3437–3452. [Google Scholar] [CrossRef]
- Liepelt, S.; Cheddadi, R.; de Beaulieu, J.L.; Fady, B.; Gömöry, D.; Hussendörfer, E.; Konnert, M.; Litt, T.; Longauer, R.; Terhürne-Berson, R.; et al. Postglacial range expansion and its genetic imprints in Abies alba (Mill.)—A synthesis from palaeobotanic and genetic data. Rev. Palaeobot. Palynol. 2009, 153, 139–149. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Aravanopoulos, F.A.; Argiriou, A.; Kalivas, A.; Tsaftaris, A. Is the genetic diversity of small scattered forest tree populations at the southern limits of their range more prone to stochastic events? A wild cherry case study by microsatellite-based markers. Tree Genet. Genomes 2011, 7, 1299–1313. [Google Scholar] [CrossRef]
- Cornille, A.; Giraud, T.; Bellard, C.; Tellier, A.; Le Cam, B.; Smulders, M.J.M.; Kleinschmit, J.; Roldan-Ruiz, I.; Gladieux, P. Postglacial recolonization history of the European crabapple (Malus sylvestris Mill.), a wild contributor to the domesticated apple. Mol. Ecol. 2013, 22, 2249–2263. [Google Scholar] [CrossRef]
- King, A.; Ferris, C. Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Mol. Ecol. 1998, 7, 1151–1161. [Google Scholar] [CrossRef]
- Douda, J.; Doudova, J.; Drašnarová, A.; Kuneš, P.; Hadincová, V.; Krak, K.; Zákravský, P.; Mandak, B. Migration patterns of subgenus Alnus in Europe since the Last Glacial Maximum: A systematic review. PLoS ONE 2014, 9, 88709. [Google Scholar] [CrossRef]
- Mejnartowicz, L. Genetic variation within and among naturally regenerating populations of alder (Alnus glutinosa). Acta Soc. Bot. Ploniae 2008, 77, 105–110. [Google Scholar] [CrossRef]
- Palstra, F.P.; Ruzzante, D.E. Genetic estimates of contemporary effective population size: What can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 2008, 17, 3428–3447. [Google Scholar] [CrossRef] [PubMed]
- Koskela, J.; Lefèvre, F.; Schueler, S.; Kraigher, H.; Olrik, D.C.; Hubert, J.; Longauer, R.; Bozzano, M.; Yrjänä, L.; Alizoti, P.; et al. Translating conservation genetics into management: Pan-European minimum requirements for dynamic conservation units of forest tree genetic diversity. Biol Conserv, 2013, 157, 39–49. [Google Scholar] [CrossRef]
- De Vries, S.M.G.; Alan, M.; Bozzano, M.; Burianek, V.; Collin, E.; Cottrell, J.; Ivankovic, M.; Kelleher, C.T.; Koskela, J.; Rotach, P.; et al. Pan-European Strategy for Genetic Conservation of Forest Trees and Establishment of a Core Network of Dynamic Conservation Units; European Forest Genetic Resources Programme (EUFORGEN), Bioversity International: Rome, Italy, 2015; p. xii + 40. [Google Scholar]
- Silviculture. In Lithuanian Statistical Yearbook of Forestry; State Forestry Srvice: Kaunas, Lithuania, 2021; pp. 105–115.
- Capon, S.J.; Chambers, L.E.; Mac Nally, R.; Naiman, R.J.; Davies, P.; Marshall, N.; Pittock, J.; Reid, M.; Capon, T.; Douglas, M.; et al. Riparian ecosystems in the 21st century: Hotspots for climate change adaptation? Ecosystems 2013, 16, 359–381. [Google Scholar] [CrossRef]
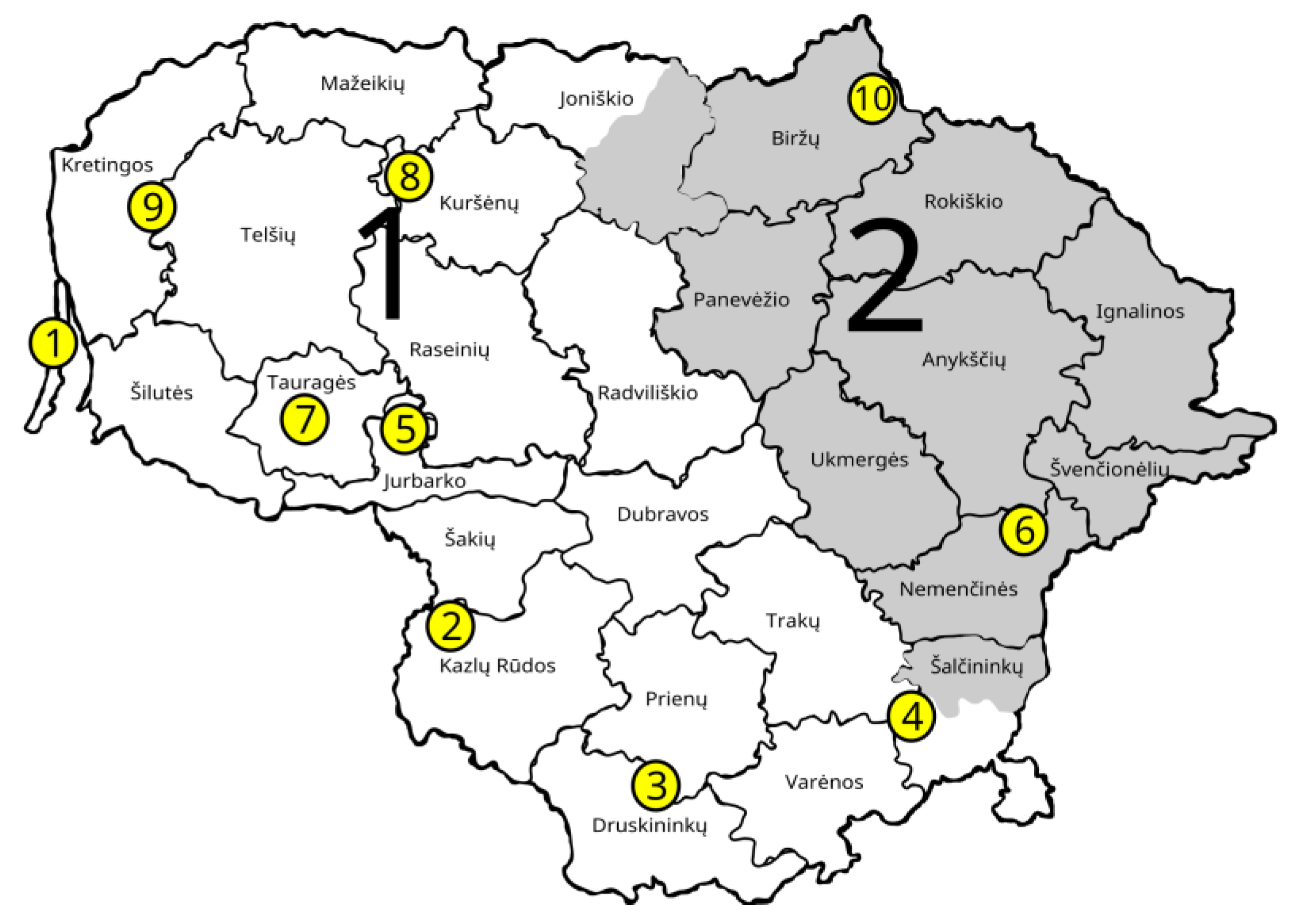

| Population Abbrev. | GCU Code | Full Name of GCU | No of Trees Sampled; Total (Mature, Juvenile) | Regional Division of State Forest Enterprise and Forest District | Species Composition in the GCU * | Coordinates |
|---|---|---|---|---|---|---|
| Latitude and Longitude | ||||||
| JUO | 35JSM037 | Juodkrantė seed stand | 58 (28, 30) | Kretinga r.d., Juodkrantė | 100% A. glutinosa | 55°36′30.8988″ 21°7′54.8004″ |
| VIL | 33JGD008 | Žalioji forest black alder genetic reserve | 60 (30, 30) | Kazlų Rūda r.d., Vilkaviškis | 100% A. glutinosa | 54°43′7.284″ 22°57′25.0914″ |
| LEI | 03JSM049 | Leipalingis seed stand | 60 (30, 30) | Veisiejai r.d., Leipalingis | 90% A. glutinosa, 10% B. pendula | 54°10′37.704″ 23°48′10.0074″ |
| GIR | 32JSM040 | Giria seed stand | 60 (30, 30) | Šalčininkai r.d., Girios | 100% A. glutinosa | 54°24′44.17142″ 5°3′15.552″ |
| SIM | 17JGD019 | Šimkaičiai forest black alder genetic reserve | 60 (30, 30) | Raseiniai r.d., Šimkaičiai | 80% A. glutinosa, 10% B. pendula, 10% P. abies | 55°11′40.416″ 22°50′9.672″ |
| PUR | 31JSM030 | Purviniškės seed stand | 60 (30, 30) | Nemenčinės r.d., Purviniškės | 100% A. glutinosa | 55°0′55.224″ 25°37′39.108″ |
| BAT | 18JGD018 | Bijotai forest black alder genetic reserve | 60 (30, 30) | Tauragė r.d., Batakiai | 100% A. glutinosa | 55°20′9.564″ 22°28′23.232″ |
| PAZ | 53JGD005/006 | Tryškiai forest black alder genetic reserve | 60 (30, 30) | Kuršėnai r.d., Pažiužmėliai | 100% A. glutinosa | 56°3′28.9434″ 22°43′11.8914″ |
| MIK | 60JSM044 | Mikoliškės seed stand | 58(28, 30) | Kretinga r.d., Mikoliškės | 100% A. glutinosa | 55°47′57.804″ 21°29′43.8354″ |
| SPA | 01JGD001 | Biržai Forest black alder genetic reserve | 61 (31, 30) | Biržai r.d., Spalviškės | 90% A. glutinosa, 10% B. pendula | 56°15′38.1594″ 24°58′55.308″ |
| Locus | Repeat Motif Size in bp | No of Alleles | Na | Ne | Allele Interval in bp | Most Common Allele and Its Frequency | The Frequency of Null Alleles | Ho | He | F | Primer Source Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A2 | 2 | 7 | 3.50 ± 0.20 | 1.67 ± 0.08 | 132–148 | 144 (0.70) | −0.1169 | 0.53 ± 0.06 | 0.36 ± 0.03 | 0.00 ± 0.07 | [40] |
| A22 | 2 | 15 | 8.55 ± 0.25 | 5.24 ± 0.22 | 151–181 | 165 (0.23) | 0.0378 | 0.78 ± 0.03 | 0.80 ± 0.02 | 0.03 ± 0.03 | [41] |
| A10 | 2 | 12 | 6.60 ± 0.37 | 3.96 ± 0.23 | 107–133 | 119 (0.29) | 0.0256 | 0.75 ± 0.03 | 0.73 ± 0.02 | −0.02 ± 0.02 | [42] |
| A35 | 2 | 16 | 9.95 ± 0.30 | 5.19 ± 0.25 | 216–242 | 228 (0.33) | 0.0943 | 0.67 ± 0.02 | 0.80 ± 0.01 | 0.16 ± 0.02 | [43] |
| A38 | 3 | 15 | 8.95 ± 0.33 | 2.86 ± 0.15 | 97–151 | 121 (0.54) | −0.0534 | 0.72 ± 0.03 | 0.63 ± 0.02 | −0.13 ± 0.02 | [43] |
| A7 | 2 | 13 | 6.10 ± 0.29 | 2.49 ± 0.22 | 168–200 | 174 (0.54) | 0.1276 | 0.52 ± 0.05 | 0.53 ± 0.04 | −0.01 ± 0.06 | [44] |
| A37 | 2 | 17 | 9.30 ± 0.28 | 4.81 ± 0.22 | 239–275 | 257 (0.29) | 0.0697 | 0.71 ± 0.03 | 0.78 ± 0.01 | 0.10 ± 0.04 | [45] |
| A26 | 2 | 19 | 11.60 ± 0.27 | 6.14 ± 0.21 | 341–377 | 353 (0.26) | 0.0555 | 0.75 ± 0.02 | 0.83 ± 0.01 | 0.10 ± 0.02 | [41] |
| Ag30 | 2 | 15 | 9.20 ± 0.31 | 5.00 ± 0.18 | 76–114 | 100 (0.20) | −0.0415 | 0.93 ± 0.01 | 0.80 ± 0.01 | −0.17 ± 0.02 | [24] |
| Ag05 | 2 | 16 | 9.55 ± 0.25 | 6.45 ± 0.23 | 136–168 | 154 (0.17) | 0.0324 | 0.81 ± 0.03 | 0.84 ± 0.01 | 0.04 ± 0.03 | [24] |
| Ag10 | 2 | 16 | 6.60 ± 0.37 | 1.75 ± 0.16 | 205–249 | 217 (0.76) | 0.0154 | 0.40 ± 0.03 | 0.38 ± 0.03 | −0.04 ± 0.02 | [24] |
| Ag14 | 2 | 31 | 12.45 ± 0.45 | 6.18 ± 0.36 | 271–341 | 289 (0.26) | 0.1504 | 0.60 ± 0.03 | 0.83 ± 0.01 | 0.27 ± 0.03 | [24] |
| Ag27 | 2 | 15 | 7.10 ± 0.32 | 3.10 ± 0.14 | 74–112 | 102 (0.47) | −0.0097 | 0.72 ± 0.04 | 0.66 ± 0.02 | −0.07 ± 0.04 | [24] |
| Ag35 | 2 | 23 | 13.25 ± 0.42 | 6.83 ± 0.34 | 164–214 | 174 (0.19) | 0.0141 | 0.86 ± 0.01 | 0.84 ± 0.01 | −0.02 ± 0.02 | [24] |
| Ag13 | 2 | 17 | 11.50 ± 0.42 | 7.48 ± 0.36 | 243–283 | 273 (0.17) | 0.0133 | 0.87 ± 0.02 | 0.85 ± 0.02 | −0.02 ± 0.02 | [24] |
| Ag25 | 2 | 23 | 9.10 ± 0.36 | 3.82 ± 0.21 | 73–131 | 103 (0.24) | 0.017 | 0.80 ± 0.03 | 0.73 ± 0.01 | −0.10 ± 0.05 | [24] |
| Ag09 | 2 | 21 | 8.35 ± 0.26 | 5.32 ± 0.17 | 200–264 | 244 (0.26) | 0.0042 | 0.81 ± 0.02 | 0.81 ± 0.01 | −0.01 ± 0.02 | [24] |
| Ag20 | 2 | 19 | 6.75 ± 0.63 | 2.94 ± 0.39 | 283–333 | 305 (0.56) | 0.1825 | 0.45 ± 0.05 | 0.52 ± 0.06 | 0.09 ± 0.04 | [24] |
| Ag01 | 2 | 12 | 7.30 ± 0.18 | 3.90 ± 0.16 | 126–188 | 130 (0.36) | −0.0128 | 0.80 ± 0.03 | 0.73 ± 0.02 | −0.09 ± 0.03 | [24] |
| Ag23 | 2 | 9 | 3.50 ± 0.12 | 2.31 ± 0.06 | 336–370 | 356 (0.54) | 0.1042 | 0.45 ± 0.03 | 0.56 ± 0.01 | 0.20 ± 0.05 | [24] |
| Population Cohort | Ne | Ar | Ho | He | Rare Alleles No/Freq. | Private Alleles No/Freq. | F | Effective Pop Size (95% CI) | LGP |
|---|---|---|---|---|---|---|---|---|---|
| JUO-J | 3.89 ± 0.41 | 5.68 ± 0.36 (7.27 ± 0.45) | 0.58 ± 0.05 | 0.68 ± 0.03 | 87/4.50 ± 0.33 | 14/0.75 ± 0.19 | 0.15 ± 0.05 | 9.6 (8.8) | 4.56 ± 0.37 |
| JUO-M | 4.00 ± 0.42 | 5.09 ± 0.37 (6.76 ± 0.54) | 0.63 ± 0.05 | 0.66 ± 0.05 | 57/4.85 ± 0.45 | 3/0.15 ± 0.08 | 0.03 ± 0.05 | 740.3 (176.8) | 3.70 ± 0.41 |
| VIL-J | 4.83 ± 0.49 | 5.51 ± 0.36 (7.85 ± 0.51) | 0.81 ± 0.04 | 0.74 ± 0.03 | 90/5.25 ± 0.44 | 6/0.30 ± 0.13 | −0.11 ± 0.04 | 583.9 (206.5) | 4.42 ± 0.40 |
| VIL-M | 4.01 ± 0.44 | 4.84 ± 0.35 (6.88 ± 0.55) | 0.65 ± 0.05 | 0.67 ± 0.04 | 76/4.85 ± 0.42 | 2/0.10 ± 0.07 | 0.01 ± 0.05 | 1515 (243.3) | 3.99 ± 0.44 |
| LEI-J | 4.81 ± 0.48 | 5.09 ± 0.32 (8.15 ± 0.52) | 0.74 ± 0.04 | 0.74 ± 0.04 | 106/5.10 ± 0.43 | 4/0.20 ± 0.09 | −0.02 ± 0.04 | 97.4 (73.8) | 4.89 ± 0.47 |
| LEI-M | 4.34 ± 0.50 | 4.95 ± 0.38 (7.41 ± 0.56) | 0.68 ± 0.05 | 0.70 ± 0.04 | 94/4.80 ± 0.43 | 3/0.15 ± 0.08 | 0.01 ± 0.06 | 174.7 (110.2) | 4.61 ± 0.40 |
| GIR-J | 4.69 ± 0.44 | 5.68 ± 0.36 (7.66 ± 0.50) | 0.75 ± 0.05 | 0.73 ± 0.04 | 76/5.40 ± 0.45 | 3/0.15 ± 0.08 | −0.04 ± 0.05 | 248.6 (133.8) | 4.16 ± 0.53 |
| GIR-M | 3.88 ± 0.40 | 5.22 ± 0.39 (6.46 ± 0.49) | 0.67 ± 0.05 | 0.67 ± 0.04 | 57/5.00 ± 0.39 | 0 | −0.02 ± 0.06 | 5731.9 (241.8) | 3.47 ± 0.36 |
| SIM-J | 4.86 ± 0.44 | 5.39 ± 0.31 (7.81 ± 0.48) | 0.75 ± 0.04 | 0.75 ± 0.03 | 82/5.60 ± 0.40 | 2/0.10 ± 0.07 | −0.01 ± 0.03 | 190.1 (116.0) | 4.19 ± 0.37 |
| SIM-M | 4.39 ± 0.53 | 5.10 ± 0.36 (7.45 ± 0.57) | 0.67 ± 0.05 | 0.68 ± 0.04 | 76/5.35 ± 0.46 | 0 | 0.01 ± 0.03 | 221.3 (122.9) | 4.06 ± 0.39 |
| PUR-J | 4.30 ± 0.41 | 5.25 ± 0.35 (7.41 ± 0.43) | 0.75 ± 0.05 | 0.71 ± 0.04 | 78/5.45 ± 0.39 | 3/0.15 ± 0.08 | −0.06 ± 0.04 | 109.7 (77.0) | 4.05 ± 0.40 |
| PUR-M | 4.11 ± 0.46 | 4.92 ± 0.34 (7.01 ± 0.51) | 0.65 ± 0.05 | 0.66 ± 0.05 | 82/4.80 ± 0.50 | 1/0.05 ± 0.05 | 0.01 ± 0.04 | 184.0 (107.9) | 4.04 ± 0.39 |
| BAT-J | 4.98 ± 0.48 | 5.34 ± 0.33 (7.81 ± 0.49) | 0.70 ± 0.04 | 0.74 ± 0.04 | 76/5.95 ± 0.46 | 1/0.05 ± 0.05 | 0.05 ± 0.03 | 93.4 (68.2) | 3.77 ± 0.40 |
| BAT-M | 4.18 ± 0.46 | 5.04 ± 0.38 (7.02 ± 0.51) | 0.71 ± 0.04 | 0.69 ± 0.04 | 68/5.05 ± 0.50 | 0 | −0.05 ± 0.04 | 760.7 (200.1) | 3.87 ± 0.43 |
| PAZ-J | 4.40 ± 0.39 | 5.64 ± 0.34 (7.13 ± 0.51) | 0.72 ± 0.05 | 0.71 ± 0.04 | 72/5.10 ± 0.36 | 2/0.10 ± 0.07 | −0.02 ± 0.03 | 83.5 (62.4) | 3.85 ± 0.41 |
| PAZ-M | 3.93 ± 0.40 | 5.34 ± 0.40 (6.75 ± 0.46) | 0.66 ± 0.05 | 0.68 ± 0.04 | 70/4.85 ± 0.43 | 3/0.15 ± 0.08 | 0.02 ± 0.06 | 720.8 (194.4) | 3.87 ± 0.33 |
| MIK-J | 4.29 ± 0.38 | 5.62 ± 0.36 (7.56 ± 0.47) | 0.65 ± 0.04 | 0.72 ± 0.03 | 86/5.05 ± 0.30 | 1/0.05 ± 0.05 | 0.08 ± 0.04 | 48.1 (39.6) | 4.41 ± 0.42 |
| MIK-M | 4.02 ± 0.40 | 5.43 ± 0.42 (7.07 ± 0.50) | 0.66 ± 0.05 | 0.68 ± 0.04 | 71/4.65 ± 0.37 | 3/0.15 ± 0.11 | 0.002 ± 0.05 | 400.7 (155.9) | 4.18 ± 0.33 |
| SPA-J | 4.89 ± 0.49 | 5.61 ± 0.35 (7.71 ± 0.47) | 0.71 ± 0.05 | 0.73 ± 0.04 | 70/5.40 ± 0.44 | 2/0.10 ± 0.07 | 0.02 ± 0.04 | 75.8 (57.4) | 3.71 ± 0.44 |
| SPA-M | 4.66 ± 0.51 | 4.98 ± 0.38 (7.46 ± 0.63) | 0.76 ± 0.05 | 0.71 ± 0.04 | 76/4.80 ± 0.40 | 2/0.10 ± 07 | −0.09 ± 0.05 | 355.6 (170.8) | 3.93 ± 0.47 |
| Mean/total | 4.37 ± 0.10 | 5.77 ± 0.33 (7.33 ± 0.12) | 0.70 ± 0.01 | 0.70 ± 0.01 | 1550 (77.5) | 55 (2.75) | −0.001 ± 0.01 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verbylaitė, R.; Aravanopoulos, F.A.; Baliuckas, V.; Juškauskaitė, A. Genetic Monitoring of Alnus glutinosa Natural Populations Using Two Generation Cohorts. Forests 2023, 14, 330. https://doi.org/10.3390/f14020330
Verbylaitė R, Aravanopoulos FA, Baliuckas V, Juškauskaitė A. Genetic Monitoring of Alnus glutinosa Natural Populations Using Two Generation Cohorts. Forests. 2023; 14(2):330. https://doi.org/10.3390/f14020330
Chicago/Turabian StyleVerbylaitė, Rita, Filippos A. Aravanopoulos, Virgilijus Baliuckas, and Aušra Juškauskaitė. 2023. "Genetic Monitoring of Alnus glutinosa Natural Populations Using Two Generation Cohorts" Forests 14, no. 2: 330. https://doi.org/10.3390/f14020330
APA StyleVerbylaitė, R., Aravanopoulos, F. A., Baliuckas, V., & Juškauskaitė, A. (2023). Genetic Monitoring of Alnus glutinosa Natural Populations Using Two Generation Cohorts. Forests, 14(2), 330. https://doi.org/10.3390/f14020330






