A Novel Simplified Protocol for Pre-Processing Whole Wood Samples for Stable Isotope Analysis in Tree Rings
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Material and Pre-Treatment
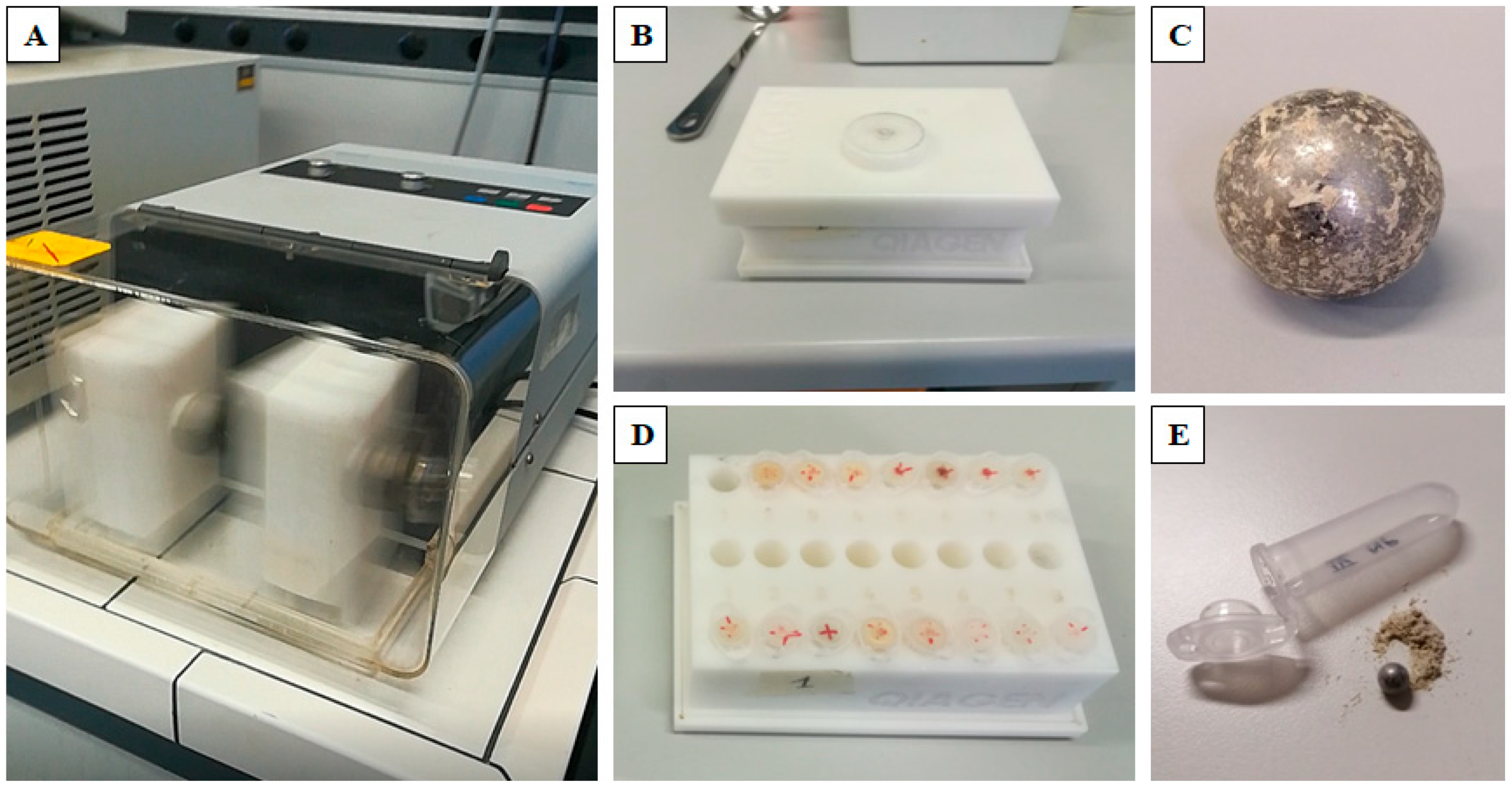
2.2. Standard Pre-Processing Protocol
2.3. New Pre-Processing Protocol
2.4. Analysis of Wood Particle Size from Different Protocols
2.5. Statistical Analyses
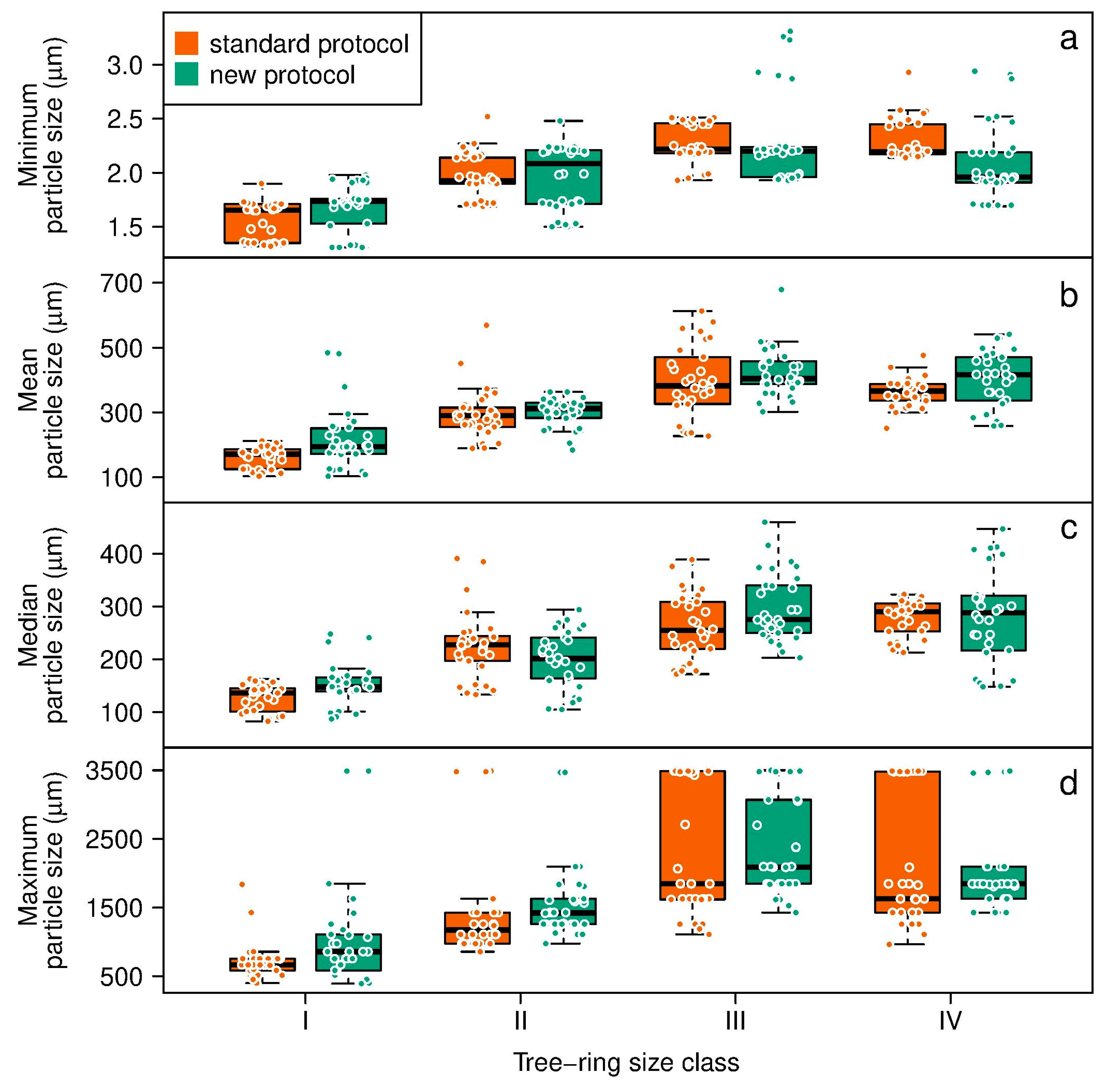
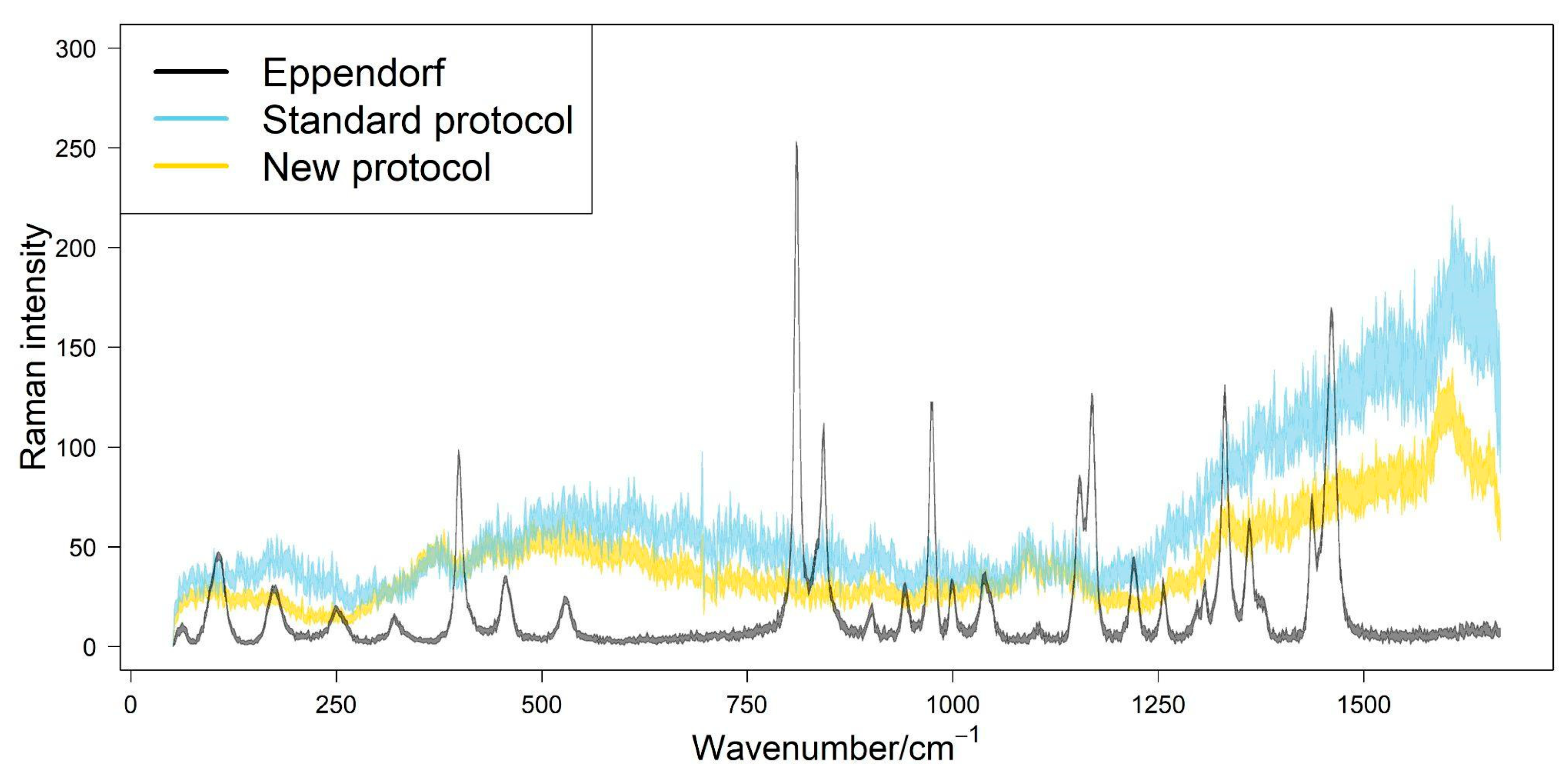
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.H.; et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef]
- Senf, C.; Buras, A.; Zang, C.S.; Rammig, A.; Seidl, R. Excess forest mortality is consistently linked to drought across Europe. Nat. Commun. 2020, 11, 6200. [Google Scholar] [CrossRef] [PubMed]
- Anderegg, W.R.; Trugman, A.T.; Badgley, G.; Konings, A.G.; Shaw, J. Divergent forest sensitivity to repeated extreme droughts. Nat. Clim. Chang. 2020, 10, 1091–1095. [Google Scholar] [CrossRef]
- Gazol, A.; Camarero, J.J.; Vicente-Serrano, S.M.; Sánchez-Salguero, R.; Gutiérrez, E.; de Luis, M.; Sangüesa-Barreda, G.; Novak, K.; Rozas, V.; Tíscar, P.A.; et al. Forest resilience to drought varies across biomes. Glob. Change Biol. 2018, 24, 2143–2158. [Google Scholar] [CrossRef]
- Gentilesca, T.; Camarero, J.J.; Colangelo, M.; Nolè, A.; Ripullone, F. Drought-induced oak decline in the western Mediterranean region: An overview on current evidences, mechanisms and management options to improve forest resilience. Iforest Biogeosci. For. 2017, 10, 796–806. [Google Scholar] [CrossRef]
- Babst, F.; Bouriaud, O.; Poulter, B.; Trouet, V.; Girardin, M.P.; Frank, D.C. Twentieth century redistribution in climatic drivers of global tree growth. Sci. Adv. 2019, 5, eaat4313. [Google Scholar] [CrossRef] [PubMed]
- Batllori, E.; Lloret, F.; Aakala, T.; Anderegg, W.R.; Aynekulu, E.; Bendixsen, D.P.; Bentouati, A.; Bigler, C.; Burk, C.J.; Camarero, J.J.; et al. Forest and woodland replacement patterns following drought-related mortality. Proc. Natl. Acad. Sci. USA 2020, 117, 29720–29729. [Google Scholar] [CrossRef]
- McDowell, N.G.; Allen, C.D.; Anderson-Teixeira, K.; Aukema, B.H.; Bond-Lamberty, B.; Chini, L.; Clark, J.S.; Dietze, M.; Grossiord, C.; Hanbury-Brown, A.; et al. Pervasive shifts in forest dynamics in a changing world. Science 2020, 368, eaaz9463. [Google Scholar] [CrossRef]
- Colangelo, M.; Camarero, J.J.; Battipaglia, G.; Borghetti, M.; De Micco, V.; Gentilesca, T.; Ripullone, F. A multi-proxy assessment of dieback causes in a Mediterranean oak species. Tree Physiol. 2017, 37, 617–631. [Google Scholar] [CrossRef]
- Puchi, P.F.; Camarero, J.J.; Battipaglia, G.; Carrer, M. Retrospective analysis of wood anatomical traits and tree-ring isotopes suggests site-specific mechanisms triggering Araucaria araucana drought-induced dieback. Glob. Change Biol. 2021, 27, 6394–6408. [Google Scholar] [CrossRef]
- Siegwolf, R.T.; Brooks, J.R.; Roden, J.; Saurer, M. Stable Isotopes in Tree Rings: Inferring Physiological, Climatic and Environmental Responses; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Camarero, J.J.; Colangelo, M.; Rodríguez-González, P.M. Tree growth, wood anatomy and carbon and oxygen isotopes responses to drought in Mediterranean riparian forests. For. Ecol. Manag. 2023, 529, 120710. [Google Scholar] [CrossRef]
- Castagneri, D.; Battipaglia, G.; von Arx, G.; Pacheco, A.; Carrer, M. Tree-ring anatomy and carbon isotope ratio show both direct and legacy effects of climate on bimodal xylem formation in Pinus pinea. Tree Physiol. 2018, 38, 1098–1109. [Google Scholar] [CrossRef]
- Voltas, J.; Camarero, J.J.; Carulla, D.; Aguilera, M.; Ortiz, A.; Ferrio, J.P. A retrospective, dual-isotope approach reveals individual predispositions to winter-drought induced tree dieback in the southernmost distribution limit of Scots pine. Plant Cell Environ. 2013, 36, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Dobbertin, M. Tree growth as indicator of tree vitality and of tree reaction to environmental stress: A review. Eur. J. For. Res. 2005, 124, 319–333. [Google Scholar] [CrossRef]
- Pellizzari, E.; Camarero, J.J.; Gazol, A.; Sangüesa-Barreda, G.; Carrer, M. Wood anatomy and carbon-isotope discrimination support long-term hydraulic deterioration as a major cause of drought-induced dieback. Glob. Change Biol. 2016, 22, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Belmecheri, S.; Wright, W.E.; Szejner, P. Sample Collection and Preparation for Annual and Intra-annual Tree-Ring Isotope Chronologies. In Stable Isotopes in Tree Rings: Inferring Physiological, Climatic and Environmental Responses; Siegwolf, R.T.W., Brooks, J.R., Roden, J., Saurer, M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 103–134. ISBN 978-3-030-92697-7. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.S.; Sarkar, D.; R Core Team. Nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-157. 2022. Available online: https://CRAN.R-project.org/package=nlme (accessed on 9 March 2023).
- Fox, J.; Weisberg, S. An {R} Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 9 March 2023).
- Schollaen, K.; Baschek, H.; Heinrich, I.; Slotta, F.; Pauly, M.; Helle, G. A guideline for sample preparation in modern tree-ring stable isotope research. Dendrochronologia 2017, 44, 133–145. [Google Scholar] [CrossRef]
- Gessler, A.; Cailleret, M.; Joseph, J.; Schönbeck, L.; Schaub, M.; Lehmann, M.; Treydte, K.; Rigling, A.; Timofeeva, G.; Saurer, M. Drought induced tree mortality–a tree-ring isotope based conceptual model to assess mechanisms and predispositions. New Phytol. 2018, 219, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Shestakova, T.A.; Martínez-Sancho, E. Stories hidden in tree rings: A review on the application of stable carbon isotopes to dendrosciences. Dendrochronologia 2021, 65, 125789. [Google Scholar] [CrossRef]
- Belmecheri, S.; Wright, W.E.; Szejner, P.; Morino, K.A.; Monson, R.K. Carbon and oxygen isotope fractionations in tree rings reveal interactions between cambial phenology and seasonal climate. Plant Cell Environ. 2018, 41, 2758–2772. [Google Scholar] [CrossRef]
- Monson, R.K.; Szejner, P.; Belmecheri, S.; Morino, K.A.; Wright, W.E. Finding the seasons in tree ring stable isotope ratios. Am. J. Bot. 2018, 105, 819–821. [Google Scholar] [CrossRef]
- Camarero, J.J.; Olano, J.M.; Parras, A. Plastic bimodal xylogenesis in conifers from continental Mediterranean climates. New Phytol. 2010, 185, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Camarero, J.J.; Carrer, M. Linking wood anatomy and xylogenesis allows pinpointing of climate and drought influences on growth of coexisting conifers in continental Mediterranean climate. Tree Physiol. 2016, 36, 502–512. [Google Scholar] [CrossRef] [PubMed]
- McCarroll, D.; Pawellek, F. Stable carbon isotope ratios of latewood cellulose in Pinus sylvestris from northern Fin-land: Variability and signal-strength. Holocene 1998, 8, 675–684. [Google Scholar] [CrossRef]
- Ponton, S.; Dupouey, J.L.; Bréda, N.; Feuillat, F.; Bodénès, C.; Dreyer, E. Carbon isotope discrimination and wood anatomy variations in mixed stands of Quercus robur and Quercus petraea. Plant Cell Environ. 2001, 24, 861–868. [Google Scholar] [CrossRef]
- Borella, S.; Leuenberger, M.; Saurer, M.; Siegwolf, R. Reducing uncertainties in δ13C analysis of tree rings: Pooling, milling, and cellulose extraction. J. Geophys. Res. Atmos. 1998, 103, 19519–19526. [Google Scholar] [CrossRef]
- Loader, N.J.; McCarroll, D.; Gagen, M.; Robertson, I.; Jalkanen, R. Extracting Climatic Information from Stable Isotopes in Tree Rings. Terr. Ecol. 2007, 1, 25–48. [Google Scholar] [CrossRef]
- Martínez-Sancho, E.; Slámová, L.; Morganti, S.; Grefen, C.; Carvalho, B.; Dauphin, B.; Rellstab, C.; Gugerli, F.; Opgenoorth, L.; Heer, K.; et al. The GenTree Dendroecological Collection, tree-ring and wood density data from seven tree species across Europe. Sci. Data 2020, 7, 1. [Google Scholar] [CrossRef]
- Opgenoorth, L.; Dauphin, B.; Benavides, R.; Heer, K.; Alizoti, P.; Martínez-Sancho, E.; Alía, R.; Ambrosio, O.; Audrey, A.; Auñón, F.; et al. The GenTree Platform: Growth traits and tree-level environmental data in 12 European forest tree species. Gigascience 2021, 10, giab010. [Google Scholar] [CrossRef]
- Housset, J.M.; Nadeau, S.; Isabel, N.; Depardieu, C.; Duchesne, I.; Lenz, P.; Girardin, M.P. Tree rings provide a new class of phenotypes for genetic associations that foster insights into adaptation of conifers to climate change. New Phytol. 2018, 218, 630–645. [Google Scholar] [CrossRef]
- Martínez-Sancho, E.; Rellstab, C.; Guillaume, F.; Bigler, C.; Fonti, P.; Wohlgemuth, T.; Vitasse, Y. Post-glacial re-colonization and natural selection have shaped growth responses of silver fir across Europe. Sci. Total. Environ. 2021, 779, 146393. [Google Scholar] [CrossRef]
- Avanzi, C.; Heer, K.; Büntgen, U.; Labriola, M.; Leonardi, S.; Opgenoorth, L.; Piermattei, A.; Urbinati, C.; Vendramin, G.G.; Piotti, A. Individual reproductive success in Norway spruce natural populations depends on growth rate, age and sensitivity to temperature. Heredity 2020, 124, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Avanzi, C.; Piermattei, A.; Piotti, A.; Büntgen, U.; Heer, K.; Opgenoorth, L.; Spanu, I.; Urbinati, C.; Vendramin, G.G.; Leonardi, S. Disentangling the effects of spatial proximity and genetic similarity on individual growth performances in Norway spruce natural populations. Sci. Total. Environ. 2019, 650, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Heer, K.; Behringer, D.; Piermattei, A.; Bässler, C.; Brandl, R.; Fady, B.; Jehl, H.; Liepelt, S.; Lorch, S.; Piotti, A.; et al. Linking dendroecology and association genetics in natural populations: Stress responses archived in tree rings associate with SNP genotypes in silver fir (Abies alba Mill.). Mol. Ecol. 2018, 27, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, M.; Pampuch, T.; Heer, K.; Avanzi, C.; Würth, D.G.; Trouillier, M.; Bog, M.; Wilmking, M.; Schnittler, M. Population structure and the influence of microenvironment and genetic similarity on individual growth at Alaskan white spruce treelines. Sci. Total. Environ. 2021, 798, 149267. [Google Scholar] [CrossRef] [PubMed]
| Steps of the Standard Protocol | Total Time Per 1 Run (2 Samples per Time) (Seconds) | Time per 1 Sample (Seconds) | Steps of the New Protocol | Total Time per 1 Run (24 Samples per Time) (Seconds) | Time per 1 Sample (Seconds) |
|---|---|---|---|---|---|
| Adding steel balls into grinding jars | 4.0 | 2.0 | Adding steel beads intoeach Eppendorf tube | 48.0 | 2.0 |
| Fixing and adjusting jars in the mill | 25.0 | 12.5 | Fixing and adjusting plates in the mill | 25.0 | 1.0 |
| Samples milling in grinding jars | 20.0 | 10.0 | Samples milling in plates | 60.0 | 2.5 |
| Recovering the milled material from the jarsand the steel balls | 330.0 | 165.0 | Removing steelbeads | 132.0 | 5.5 |
| Material transfer from the grinding jar to the Eppendorf tube | 90.0 | 45.0 | Not required | - | - |
| Washing of the grinding jars andthe steel balls for reuse | 99.0 | 49.5 | Washing of the steel beads for reuse | 24.0 | 1.0 |
| TIMING STANDARD PROTOCOL | 568.0 | 284.0 | TIMING NEW PROTOCOL | 289.0 | 12.0 |
| Response Variable | Fixed Effect | χ2 | Df | p (α = 0.05) | Variance Structure of the Chosen Model |
|---|---|---|---|---|---|
| Minimum | protocol | 0.241 | 1 | 0.623 | varIdent (form = ~ 1|class) |
| class | 42.272 | 3 | 3.513 × 10−9 | ||
| protocol × class | 1.639 | 3 | 0.651 | ||
| Mean | protocol | 2.137 | 1 | 0.144 | VarComb (varIdent (form = ~ 1|class), varIdent (form = ~ 1|protocol)) |
| class | 62.993 | 3 | 1.348 × 10−13 | ||
| protocol × class | 0.665 | 3 | 0.881 | ||
| Median | protocol | 0.023 | 1 | 0.879 | VarComb (varIdent (form = ~ 1|class), varIdent (form = ~ 1|protocol)) |
| class | 36.316 | 3 | 6.420 × 10−8 | ||
| protocol × class | 1.891 | 3 | 0.595 | ||
| Maximum | protocol | 0.109 | 1 | 0.741 | varIdent (form = ~ 1) |
| class | 42.564 | 3 | 3.045 × 10−9 | ||
| protocol × class | 0.783 | 3 | 0.854 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pericolo, O.; Avanzi, C.; Piotti, A.; Ripullone, F.; Nola, P. A Novel Simplified Protocol for Pre-Processing Whole Wood Samples for Stable Isotope Analysis in Tree Rings. Forests 2023, 14, 631. https://doi.org/10.3390/f14030631
Pericolo O, Avanzi C, Piotti A, Ripullone F, Nola P. A Novel Simplified Protocol for Pre-Processing Whole Wood Samples for Stable Isotope Analysis in Tree Rings. Forests. 2023; 14(3):631. https://doi.org/10.3390/f14030631
Chicago/Turabian StylePericolo, Osvaldo, Camilla Avanzi, Andrea Piotti, Francesco Ripullone, and Paola Nola. 2023. "A Novel Simplified Protocol for Pre-Processing Whole Wood Samples for Stable Isotope Analysis in Tree Rings" Forests 14, no. 3: 631. https://doi.org/10.3390/f14030631
APA StylePericolo, O., Avanzi, C., Piotti, A., Ripullone, F., & Nola, P. (2023). A Novel Simplified Protocol for Pre-Processing Whole Wood Samples for Stable Isotope Analysis in Tree Rings. Forests, 14(3), 631. https://doi.org/10.3390/f14030631






