Identification of ApbHLH1 as a Partner Interacting with ApMYB1 to Promote Anthocyanin Biosynthesis during Autumnal Leaf Coloration in Acer palmatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurement of Chlorophyll and Anthocyanin Contents in Leaves
2.3. Phylogenetic Analysis of Putative bHLH Proteins
2.4. Isolation of a Putative Target ApbHLH1 and Protein Structure Analysis
2.5. Expression of ApbHLH1 in Various Leaf Samples
2.6. Quantitative RT-PCR (qRT-PCR)
2.7. Luciferase Complementation Assay for Protein–Protein Interactions
2.8. Transient Overexpression of ApbHLH1 in Tobacco
2.9. Statistical Analysis
3. Results
3.1. Identification of ApbHLH1 According to Phylogenetic Analysis
3.2. Cloning ApbHLH1 cDNA and Analyzing Amino Acid Sequences
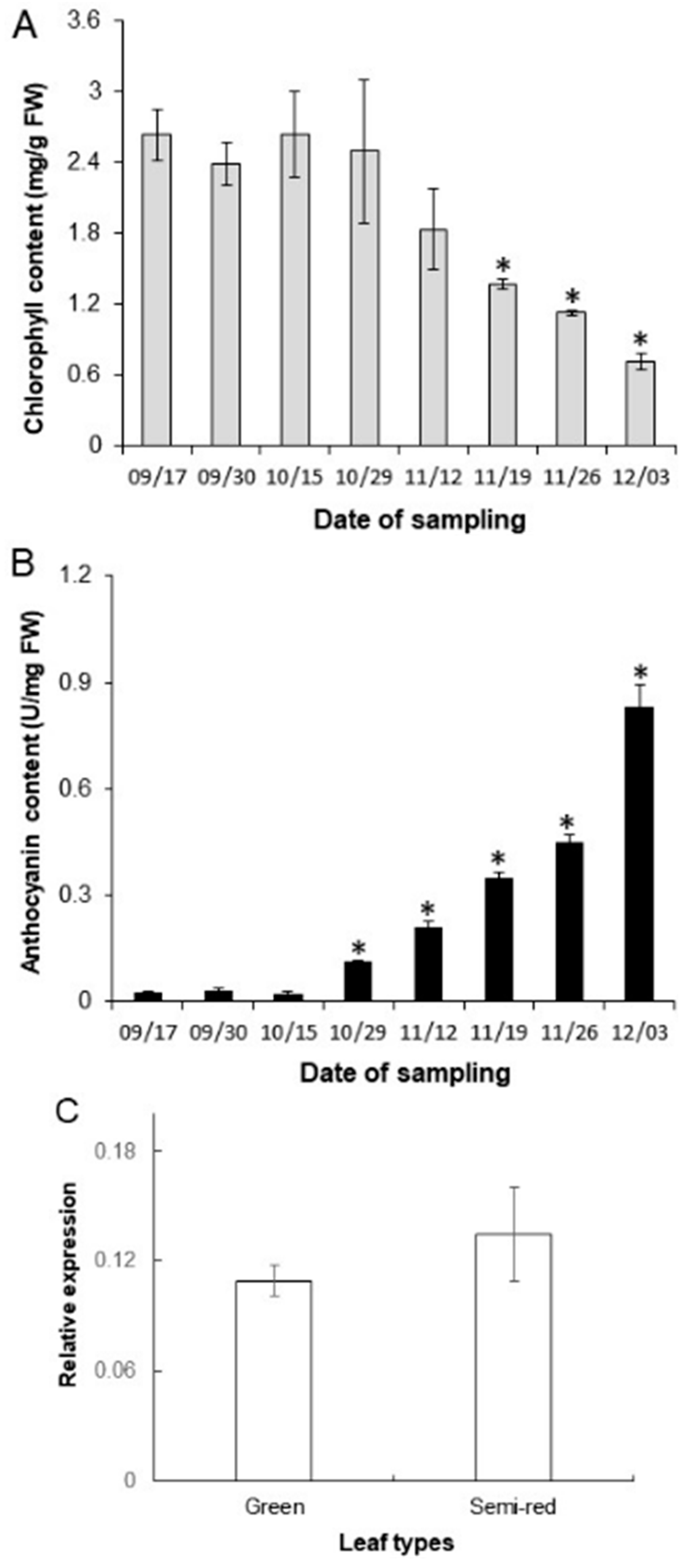
3.3. Contents of Chlorophyll and Anthocyanins and Expression Levels of ApbHLH1 during Leaf Coloration
3.4. ApbHLH1 Expression in Detached Leaves Submitted to Exogenous Treatments
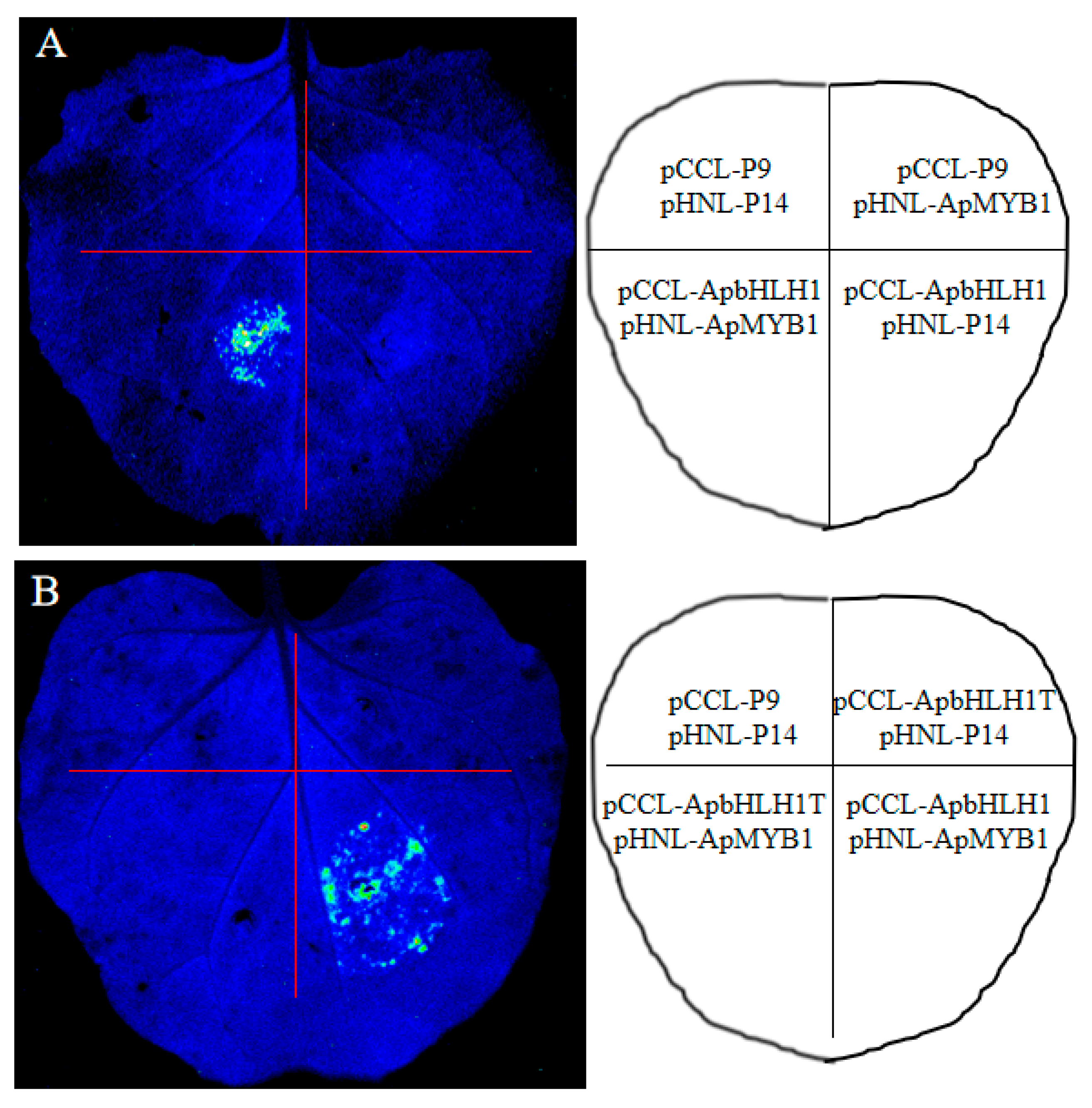
3.5. Interacting between ApbHLH1 with ApMYB1
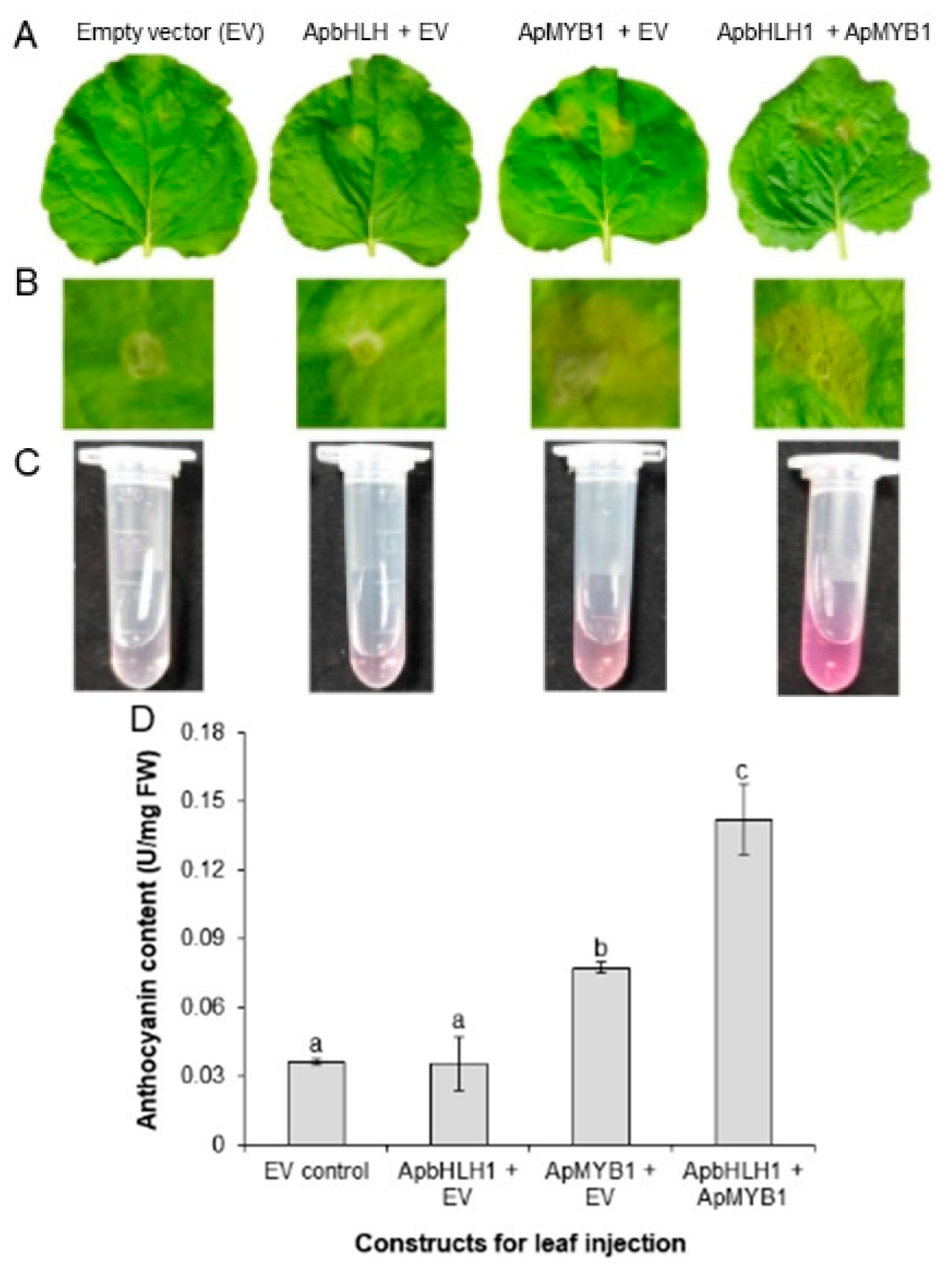
3.6. Transient Expression of ApbHLH1 in Tobacco Leaves
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Archetti, M.; Döring, T.F.; Hagen, S.B.; Hughes, N.M.; Leather, S.R.; Lee, D.W.; Lev-Yadun, S.; Manetas, Y.; Ougham, H.J.; Schaberg, P.G.; et al. Unravelling the evolution of autumn colours: An interdisciplinary approach. Trends Ecol. Evol. 2009, 24, 166–173. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, pharmacology and health benefits of anthocyanins. Phytother. Res. 2016, 30, 1265–1286. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- LaFountain, A.M.; Yuan, Y.-W. Repressors of anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Schrader, A. TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plants 2017, 6, 65. [Google Scholar] [CrossRef] [Green Version]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light induced regulation pathway of anthocyanin biosynthesis in plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef]
- Heppel, S.C.; Jaffé, F.W.; Takos, A.M.; Schellmann, S.; Rausch, T.; Walker, A.R.; Bogs, J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 2013, 82, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Zhang, B.; Chopra, D.; Schrader, A.; Hülskamp, M. Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants. J. Exp. Bot. 2019, 70, 3197–3209. [Google Scholar] [CrossRef] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.-B.; Yokoi, M.; Saito, N.; Mao, L.-S. Distribution of anthocyanins in aceraceae leaves. Biochem. Syst. Ecol. 1992, 20, 771–781. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, Q.; Yu, Y.; Feng, J.; Liu, C.; Yang, J. Leaf coloration in Acer palmatum is associated with a positive regulator ApMYB1 with potential for breeding color-leafed plants. Plants 2022, 11, 759. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.-Y.; Paik, I.; Nguyen, T.T.; Kim, J.; Oh, E. High Ambient Temperature Represses Anthocyanin Biosynthesis through Degradation of HY5. Front. Plant Sci. 2017, 8, 1787. [Google Scholar] [CrossRef] [Green Version]
- Gould, K.S.; Markham, K.R.; Smith, R.H.; Goris, J.J. Functional role of anthocyanins in the leaves of Quintinia serrata A. Cunn. J. Exp. Bot. 2000, 51, 1107–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.; Koes, R. Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Bi, G.; Zhou, J.M. Luciferase complementation assay for protein-protein interactions in plants. Curr. Protoc. Plant Biol. 2018, 3, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhou, J. Luciferase complementation assay for detecting protein interactions (in Chinese with English abstract). Chin. Bull. Bot. 2020, 55, 69–75. [Google Scholar]
- Weigel, D.; Glazebrook, J. Transformation of agrobacterium using the freeze-thaw method. Cold Spring Harb. Protoc. 2006, 2006, pdb-rot4666. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Qi, M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000, 22, 543–551. [Google Scholar] [CrossRef]
- Zhang, Y.; Wen, J.; Yau, S.S.T. Phylogenetic analysis of protein sequences based on a novel k-mer natural vector method. Genomics 2019, 111, 1298–1305. [Google Scholar] [CrossRef]
- Schmitzer, V.; Osterc, G.; Veberic, R.; Stampar, F. Correlation between chromaticity values and major anthocyanins in seven Acer palmatum Thunb. cultivars. Sci. Hortic. 2009, 119, 442–446. [Google Scholar] [CrossRef]
- Dixon, R.A.; Liu, C.; Jun, J.H. Metabolic engineering of anthocyanins and condensed tannins in plants. Curr. Opin. Biotechnol. 2013, 24, 329–335. [Google Scholar] [CrossRef]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A Conserved Network of Transcriptional Activators and Repressors Regulates Anthocyanin Pigmentation in Eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/bHLH/WD40 Complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharker, M.R.; Sukhan, Z.P.; Kim, S.C.; Lee, W.K.; Kho, K.H. Identification, characterization, and expression analysis of a serotonin receptor involved in the reproductive process of the Pacific abalone, Haliotis discus hannai. Mol. Biol. Rep. 2020, 47, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Zhang, T.; Yu, Y.; Gou, L.; Yang, J.; Xu, J.; Pi, E. Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Qu, Y.; Qi, R.; Bai, X.; Tian, G.; Wang, Y.; Wang, J.; Zhang, K. Transcriptome Analysis of the Biosynthesis of Anthocyanins in Begonia semperflorens under Low-Temperature and High-Light Conditions. Forests 2018, 9, 87. [Google Scholar] [CrossRef] [Green Version]
- Nakatsuka, A.; Yamagishi, M.; Nakano, M.; Tasaki, K.; Kobayashi, N. Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci. Hortic. 2009, 121, 84–91. [Google Scholar] [CrossRef]
- Tao, R.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-Induced Basic/Helix-Loop-Helix64 Enhances Anthocyanin Biosynthesis and Undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-Mediated Degradation in Pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, X.; Gao, J.; Guo, Y.; Huang, Z.; Du, Y. The Tomato Hoffman’s Anthocyaninless Gene Encodes a bHLH Transcription Factor Involved in Anthocyanin Biosynthesis That Is Developmentally Regulated and Induced by Low Temperatures. PLoS ONE 2016, 11, e0151067. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Zhang, Z.; Jiang, S.; Xu, H.; Wang, Y.; Feng, S.; Chen, X. Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Cult. 2016, 127, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Azuma, A.; Yakushiji, H.; Koshita, Y.; Kobayashi, S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 2012, 236, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Sun, B.; He, H.; Zhang, Y.; Tian, H.; Wang, B. Current Understanding of bHLH Transcription Factors in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, L.; Gu, H.; Cheng, D.; Guo, X.; Chen, R.; Wu, Z.; Jiang, J.; Fan, X.; Chen, J. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 2021, 11, 6863. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, P.; Chen, G.; Wu, J.; Liu, Z.; Lian, H. FvbHLH9 Functions as a Positive Regulator of Anthocyanin Biosynthesis by Forming a HY5–bHLH9 Transcription Complex in Strawberry Fruits. Plant Cell Physiol. 2020, 61, 826–837. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Sun, S.; Qiu, G.; Yang, J. Identification of ApbHLH1 as a Partner Interacting with ApMYB1 to Promote Anthocyanin Biosynthesis during Autumnal Leaf Coloration in Acer palmatum. Forests 2023, 14, 1262. https://doi.org/10.3390/f14061262
Chen L, Sun S, Qiu G, Yang J. Identification of ApbHLH1 as a Partner Interacting with ApMYB1 to Promote Anthocyanin Biosynthesis during Autumnal Leaf Coloration in Acer palmatum. Forests. 2023; 14(6):1262. https://doi.org/10.3390/f14061262
Chicago/Turabian StyleChen, Long, Sujing Sun, Gengcheng Qiu, and Jiading Yang. 2023. "Identification of ApbHLH1 as a Partner Interacting with ApMYB1 to Promote Anthocyanin Biosynthesis during Autumnal Leaf Coloration in Acer palmatum" Forests 14, no. 6: 1262. https://doi.org/10.3390/f14061262
APA StyleChen, L., Sun, S., Qiu, G., & Yang, J. (2023). Identification of ApbHLH1 as a Partner Interacting with ApMYB1 to Promote Anthocyanin Biosynthesis during Autumnal Leaf Coloration in Acer palmatum. Forests, 14(6), 1262. https://doi.org/10.3390/f14061262





