Studying the Genetic and the Epigenetic Diversity of the Endangered Species Juniperus drupacea Labill. towards Safeguarding Its Conservation in Greece
Abstract
1. Introduction
2. Materials and Methods
2.1. AFLP Procedure
2.2. MSAP Procedure
2.3. Needle Water Potential
2.4. Data Collection and Statistical Analysis
3. Results
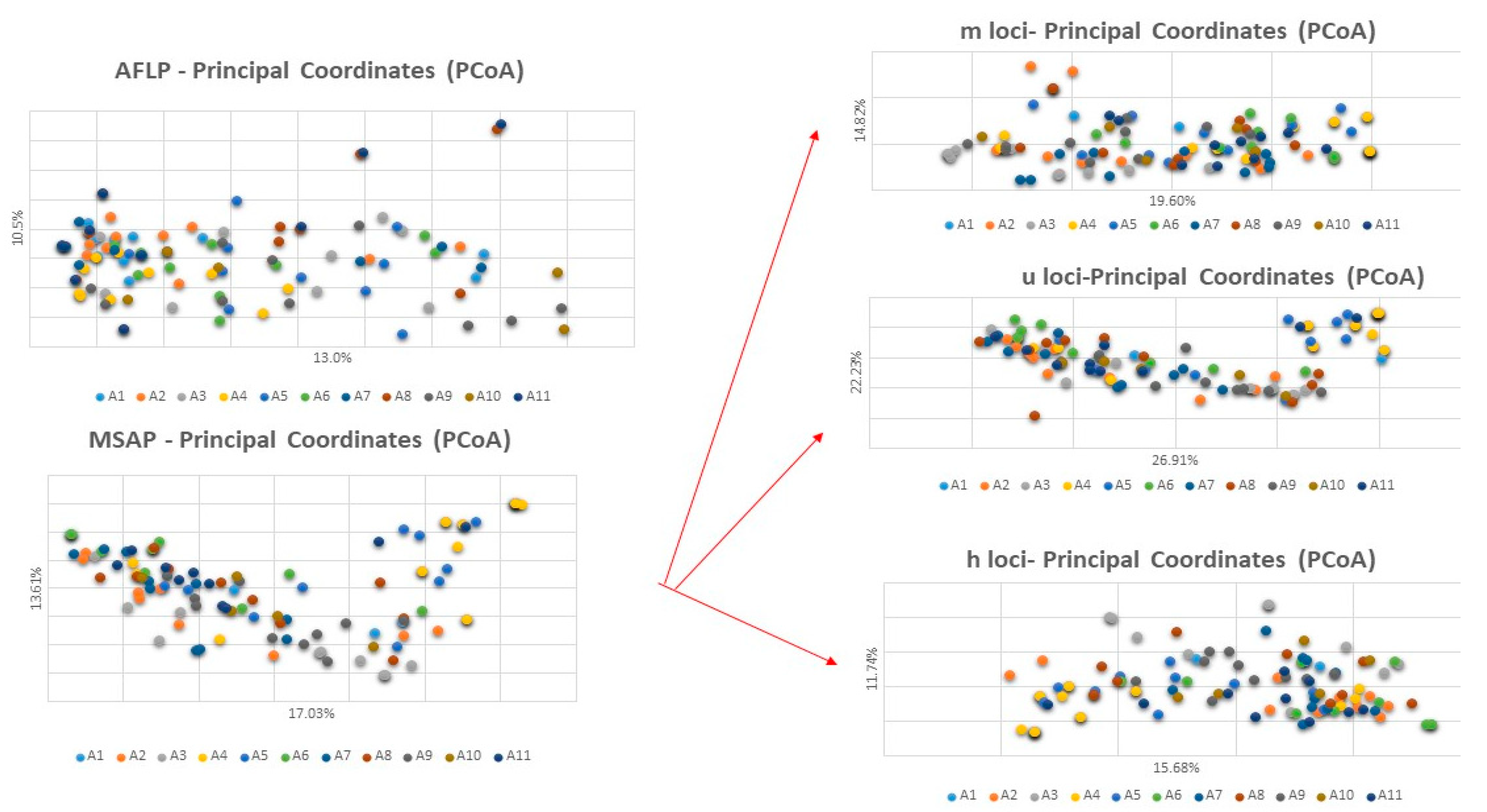
3.1. Genetic Diversity
3.2. Epigenetic Diversity
3.3. Comparison of Genetic and Epigenetic Indexes
3.4. Correlation between Geographic, Genetic, and Epigenetic Variability
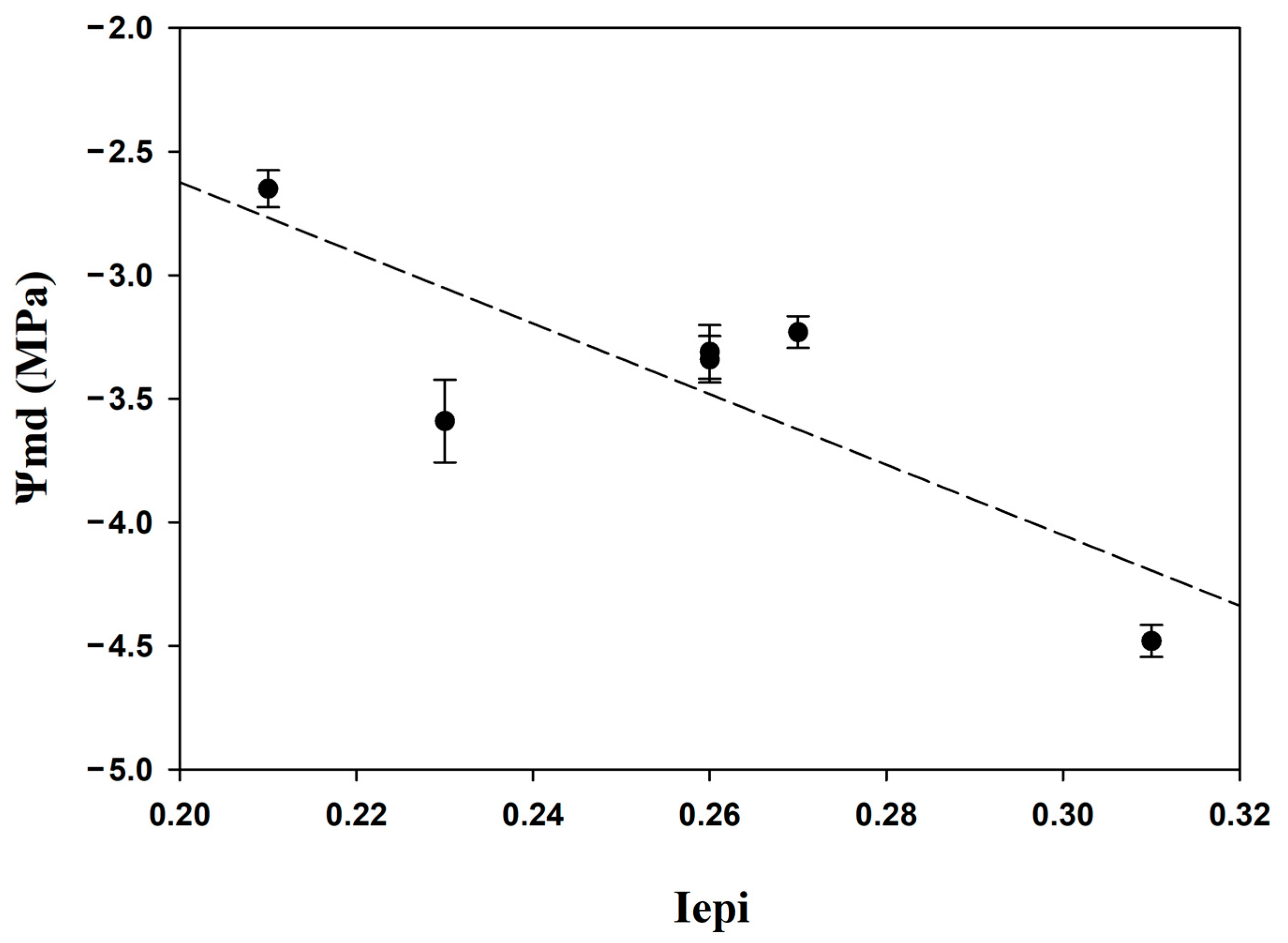
3.5. Correlation between Epigenetic Variability and Tree Water Status
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adams, R.P. Junipers of the World: The Genus Juniperus; Trafford Publishing: Bloomington, IN, USA, 2014. [Google Scholar]
- Sobierajska, K.; Boratyńska, K.; Jasińska, A.; Dering, M.; Ok, T.; Douaihy, B.; Bou Dagher-Kharrat, M.; Romo, Á.; Boratyński, A. Effect of the Aegean Sea barrier between Europe and Asia on differentiation in Juniperus drupacea (Cupressaceae). Bot. J. Linn. Soc. 2016, 180, 365–385. [Google Scholar] [CrossRef]
- Boratyński, A.; Wachowiak, W.; Dering, M.; Krystyna, B.; Katarzyna, S.; Sobierajska, K.; Jasińska, A.K.; Klimko, M.; Montserrat, J.M.; Romo, A. The biogeography and genetic relationships of Juniperus oxycedrus and related taxa from the Mediterranean and Macaronesian regions. Bot. J. Linn. Soc. 2014, 174, 637–653. [Google Scholar] [CrossRef]
- Michalczyk, I.; Sebastiani, F.; Buonamici, A.; Cremer, E.; Mengel, C.; Ziegenhagen, B.; Vendramin, G. Characterization of highly polymorphic nuclear microsatellite loci in Juniperus communis L. Mol. Ecol. Notes 2006, 6, 346–348. [Google Scholar] [CrossRef]
- Walas, Ł.; Sobierajska, K.; Ok, T.; Dönmez, A.A.; Kanoğlu, S.S.; Dagher-Kharrat, M.B.; Douaihy, B.; Romo, A.; Stephan, J.; Jasińska, A.K. Past, present, and future geographic range of an oro-Mediterranean Tertiary relict: The Juniperus drupacea case study. Reg. Environ. Chang. 2019, 19, 1507–1520. [Google Scholar] [CrossRef]
- Alfaro, R.I.; Fady, B.; Vendramin, G.G.; Dawson, I.K.; Fleming, R.A.; Sáenz-Romero, C.; Lindig-Cisneros, R.A.; Murdock, T.; Vinceti, B.; Navarro, C.M. The role of forest genetic resources in responding to biotic and abiotic factors in the context of anthropogenic climate change. For. Ecol. Manag. 2014, 333, 76–87. [Google Scholar] [CrossRef]
- Amaral, J.; Ribeyre, Z.; Vigneaud, J.; Sow, M.D.; Fichot, R.; Messier, C.; Pinto, G.; Nolet, P.; Maury, S. Advances and promises of epigenetics for forest trees. Forests 2020, 11, 976. [Google Scholar] [CrossRef]
- García-García, I.; Méndez-Cea, B.; Martín-Gálvez, D.; Seco, J.I.; Gallego, F.J.; Linares, J.C. Challenges and perspectives in the epigenetics of climate change-induced forests decline. Front. Plant Sci. 2022, 12, 3207. [Google Scholar] [CrossRef]
- Browne, L.; MacDonald, B.; Fitz-Gibbon, S.; Wright, J.W.; Sork, V.L. Genome-wide variation in DNA methylation predicts variation in leaf traits in an ecosystem-foundational oak species. Forests 2021, 12, 569. [Google Scholar] [CrossRef]
- Avramidou, E.V.; Doulis, A.G.; Aravanopoulos, F.A. Determination of epigenetic inheritance, genetic inheritance, and estimation of genome DNA methylation in a full-sib family of Cupressus sempervirens L. Gene 2015, 562, 180–187. [Google Scholar] [CrossRef]
- Raj, S.; Bräutigam, K.; Hamanishi, E.T.; Wilkins, O.; Thomas, B.R.; Schroeder, W.; Mansfield, S.D.; Plant, A.L.; Campbell, M.M. Clone history shapes Populus drought responses. Proc. Natl. Acad. Sci. USA 2011, 108, 12521–12526. [Google Scholar] [CrossRef]
- Johnsen, Ø.; Fossdal, C.G.; Nagy, N.; MØLMANN, J.; Dæhlen, O.G.; Skrøppa, T. Climatic adaptation in Picea abies progenies is affected by the temperature during zygotic embryogenesis and seed maturation. Plant Cell Environ. 2005, 28, 1090–1102. [Google Scholar] [CrossRef]
- Lira-Medeiros, C.F.; Parisod, C.; Fernandes, R.A.; Mata, C.S.; Cardoso, M.A.; Ferreira, P.C.G. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 2010, 5, e10326. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, M.; Ma, K. Whole-genome DNA methylation associated with differentially expressed genes regulated anthocyanin biosynthesis within flower color chimera of ornamental tree Prunus mume. Forests 2020, 11, 90. [Google Scholar] [CrossRef]
- Cheng, Y.-H.; Peng, X.-Y.; Yu, Y.-C.; Sun, Z.-Y.; Han, L. The effects of DNA methylation inhibition on flower development in the dioecious plant Salix viminalis. Forests 2019, 10, 173. [Google Scholar] [CrossRef]
- Saez-Laguna, E.; Guevara, M.-A.; Diaz, L.-M.; Sanchez-Gomez, D.; Collada, C.; Aranda, I.; Cervera, M.-T. Epigenetic variability in the genetically uniform forest tree species Pinus pinea L. PLoS ONE 2014, 9, e103145. [Google Scholar] [CrossRef]
- Kramer, P.J. Problems in Water Relations of Plants and Cells. In International Review Of Cytology; Elsevier: Amsterdam, The Netherlands, 1983; Volume 85, pp. 253–286. [Google Scholar]
- Richter, H.; Kikuta, S. Ecophysiology of Long-Distance Water Transport in Trees. In Trees in a Changing Environment: Ecophysiology, Adaptation, and Future Survival; Springer: Berlin/Heidelberg, Germany, 2014; pp. 99–115. [Google Scholar]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldán-Ruiz, I. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef]
- Scholander, P.F.; Bradstreet, E.D.; Hemmingsen, E.; Hammel, H. Sap Pressure in Vascular Plants: Negative hydrostatic pressure can be measured in plants. Science 1965, 148, 339–346. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol. Ecol. Resour. 2013, 13, 642–653. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Bräutigam, K.; Vining, K.J.; Lafon-Placette, C.; Fossdal, C.G.; Mirouze, M.; Marcos, J.G.; Fluch, S.; Fraga, M.F.; Guevara, M.Á.; Abarca, D. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol. Evol. 2013, 3, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Bossdorf, O.; Richards, C.L.; Pigliucci, M. Epigenetics for ecologists. Ecol. Lett. 2008, 11, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Herrera, C.M.; Bazaga, P. Untangling individual variation in natural populations: Ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Mol. Ecol. 2011, 20, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Guevara, M.Á.; Sánchez-Gómez, D.; Vélez, M.D.; de María, N.; Díaz, L.M.; Ramírez-Valiente, J.A.; Mancha, J.A.; Aranda, I.; Cervera, M.T. Epigenetic and Genetic Variability in Contrasting Latitudinal Fagus sylvatica L. Provenances. Forests 2022, 13, 1971. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Katsidi, E.C.; Avramidou, E.V.; Ganopoulos, I.; Barbas, E.; Doulis, A.G.; Triantafyllou, A.; Aravanopoulos, F.A. Genetics and epigenetics of Pinus nigra populations with differential exposure to environmental pollution. Front. Plant Sci. 2023, 14, 1178. [Google Scholar] [CrossRef]
- Rico, L.; Ogaya, R.; Barbeta, A.; Penuelas, J. Changes in DNA methylation fingerprint of Quercus ilex trees in response to experimental field drought simulating projected climate change. Plant Biol. 2014, 16, 419–427. [Google Scholar] [CrossRef]
- Schulz, B.; Eckstein, R.L.; Durka, W. Epigenetic variation reflects dynamic habitat conditions in a rare floodplain herb. Mol. Ecol. 2014, 23, 3523–3537. [Google Scholar] [CrossRef]
- Jarvis, P. The interpretation of the variations in leaf water potential and stomatal conductance found in canopies in the field. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 273, 593–610. [Google Scholar]
- Williams, L.E. Leaf water potentials of sunlit and/or shaded grapevine leaves are sensitive alternatives to stem water potential. J. Int. Sci. Vigne Vin 2012, 46, 207–219. [Google Scholar] [CrossRef]
- Alonso, C.; Pérez, R.; Bazaga, P.; Medrano, M.; Herrera, C.M. MSAP markers and global cytosine methylation in plants: A literature survey and comparative analysis for a wild-growing species. Mol. Ecol. Resour. 2016, 16, 80–90. [Google Scholar] [CrossRef]
- Medrano, M.; Herrera, C.M.; Bazaga, P. Epigenetic variation predicts regional and local intraspecific functional diversity in a perennial herb. Mol. Ecol. 2014, 23, 4926–4938. [Google Scholar] [CrossRef]
- Ioannidis, K.; Tomprou, I.; Panayiotopoulou, D.; Boutsios, S.; Daskalakou, E.N. Potential and Constraints on In Vitro Micropropagation of Juniperus drupacea Labill. Forests 2023, 14, 142. [Google Scholar] [CrossRef]
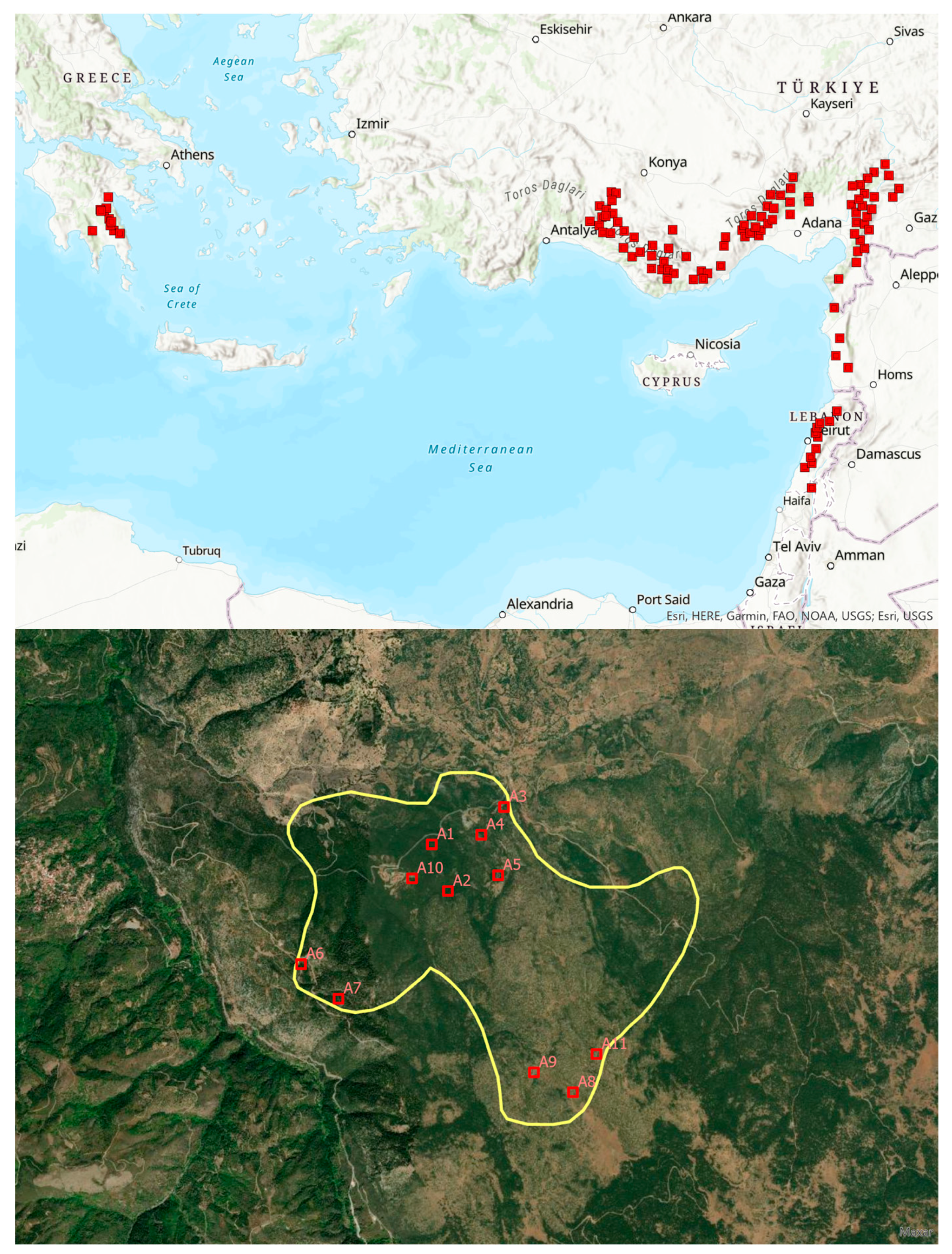

| Site Code | Latitude | Longitude | Altitude (m a.s.l.) | Number of Trees |
|---|---|---|---|---|
| A1 | 37.32904 | 22.58457 | 940 | 10 |
| A2 | 37.32679 | 22.58515 | 1046 | 10 |
| A3 | 37.33298 | 22.59284 | 949 | 10 |
| A4 | 37.33182 | 22.59195 | 964 | 10 |
| A5 | 37.32829 | 22.59287 | 1048 | 10 |
| A6 | 37.32106 | 22.57229 | 1062 | 10 |
| A7 | 37.31946 | 22.57489 | 1093 | 10 |
| A8 | 37.31065 | 22.60086 | 1270 | 10 |
| A9 | 37.31177 | 22.59781 | 1273 | 10 |
| A10 | 37.32793 | 22.58405 | 950 | 5 |
| A11 | 37.31380 | 22.60322 | 985 | 10 |
| Primer Name | 5′ to 3′ Sequence |
|---|---|
| EcoRI adapter | CTCGTAGACTGCGTACC AATTGGTACGCAGTC |
| MseI adapter | GACGATGAGTCCTGAG TACTCAGGACTCAT |
| HpaII/MspI adapter | GACGATGAGTCTCGAT CGATCGAGACTCAT |
| Pre-selective EcoRI primer | GACTGCGTACCAATTC-A |
| Pre-selective MseI primer | GATGAGTCCTGAGTAA-C |
| Pre-selective HpaII/MspI primer | ATGAGTCTCGATCGG-A |
| Selective EcoRI primers | GACTGCGTACCAATTC+ATG (FAM) GACTGCGTACCAATTC+ACT (HEX) GACTGCGTACCAATTC+AAC (ROX) GACTGCGTACCAATTC+AAG (TAMRA) |
| Selective MseI primer | GATGAGTCCTGAGTAA-CAA GATGAGTCCTGAGTAA-CAC GATGAGTCCTGAGTAA-CGT GATGAGTCCTGAGTAA-CTC |
| Selective HpaII/MspI primer | ATGAGTCTCGATCGGATC ATGAGTCTCGATCGGACT ATGAGTCTCGATCGGAAT |
| EcoRI adapter | CTCGTAGACTGCGTACC AATTGGTACGCAGTC |
| AFLP | MSAP | |||||||
|---|---|---|---|---|---|---|---|---|
| Population | P | I | He | Pepi | Iepi | Hepi | N.B. | N.P.B. |
| A1 | 47.16 | 0.15 | 0.093 | 30.26 | 0.20 | 0.068 | 486 | 9 |
| A2 | 44.77 | 0.14 | 0.088 | 48.91 | 0.31 | 0.098 | 794 | 125 |
| A3 | 53.00 | 0.17 | 0.109 | 37.43 | 0.26 | 0.088 | 621 | 59 |
| A4 | 41.53 | 0.12 | 0.076 | 32.38 | 0.21 | 0.071 | 533 | 8 |
| A5 | 60.28 | 0.20 | 0.124 | 37.37 | 0.24 | 0.080 | 606 | 48 |
| A6 | 53.00 | 0.17 | 0.106 | 32 | 0.21 | 0.078 | 533 | 32 |
| A7 | 48.43 | 0.15 | 0.095 | 37.37 | 0.26 | 0.088 | 625 | 33 |
| A8 | 53.90 | 0.17 | 0.107 | 42.36 | 0.27 | 0.089 | 683 | 74 |
| A9 | 59.85 | 0.20 | 0.125 | 40.17 | 0.26 | 0.086 | 658 | 48 |
| A10 | 40.15 | 0.17 | 0.123 | 28.51 | 0.23 | 0.077 | 480 | 22 |
| A11 | 49.12 | 0.15 | 0.096 | 37.55 | 0.25 | 0.084 | 620 | 40 |
| Mean | 50.11 | 0.16 | 0.104 | 36.75 | 0.25 | 0.083 | 603.5 | 4.40 |
| Loci/Groups | Source of Variation | d.f. | Variance Component | Total Variance (%) | Φ-Statistics (ΦST) | p Value |
|---|---|---|---|---|---|---|
| AFLP loci | Among populations | 10 | 1.88 | 1 | ||
| Within populations | 94 | 152.88 | 99 | 0.012 | <0.001 | |
| Total | 104 | 154.77 | 100 | |||
| MSAP all subepiloci | Among populations | 10 | 6.80 | 6 | ||
| Within populations | 94 | 97.28 | 94 | 0.061 | >0.001 | |
| Total | 104 | 104.09 | 100 | |||
| MSAP m-subepiloci | Among populations | 10 | 2.527 | 6 | ||
| Within populations | 94 | 38.673 | 94 | 0.038 | >0.001 | |
| Total | 104 | 41.200 | 100 | |||
| MSAP h-subepiloci | Among populations | 10 | 1.968 | 5 | ||
| Within populations | 94 | 34.796 | 95 | 0.054 | >0.001 | |
| Total | 104 | 36.764 | 100 | |||
| MSAP n-subepiloci | Among populations | 10 | 2.311 | 9 | ||
| Within populations | 94 | 23.819 | 91 | 0.088 | >0.001 | |
| Total | 104 | 26.130 | 100 |
| Population | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 0.000 | 0.000 | 0.000 | 0.0005 | 0.0001 | 0.000 | 0.000 | 0.000 | 0.0008 | 0.000 | 0.000 |
| A2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.0005 | 0.000 | 0.000 | 0.000 | 0.000 | 0.0007 | 0.000 |
| A3 | 0.000 | 0.000 | 0.000 | 0.0002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| A4 | 0.0005 | 0.0005 | 0.0002 | 0.000 | 0.0007 | 0.0006 | 0.000 | 0.0009 | 0.0006 | 0.0002 | 0.0003 |
| A5 | 0.0001 | 0.000 | 0.000 | 0.0007 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| A6 | 0.000 | 0.0001 | 0.000 | 0.0006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 |
| A7 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.0002 | 0.000 | 0.000 |
| A8 | 0.000 | 0.000 | 0.000 | 0.0009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| A9 | 0.0008 | 0.0007 | 0.000 | 0.0006 | 0.000 | 0.000 | 0.0002 | 0.000 | 0.000 | 0.000 | 0.0007 |
| A10 | 0.000 | 0.000 | 0.000 | 0.0002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| A11 | 0.000 | 0.000 | 0.000 | 0.0003 | 0.000 | 0.0002 | 0.000 | 0.000 | 0.0007 | 0.000 | 0.000 |
| u Alleles | m Alleles | h Alleles | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Subpopulation | Ne | I | He | Ne | I | He | Ne | I | He |
| A1 | 1.081 | 0.093 | 0.055 | 1.128 | 0.122 | 0.078 | 1.081 | 0.102 | 0.058 |
| A2 | 1.136 | 0.137 | 0.086 | 1.158 | 0.185 | 0.108 | 1.114 | 0.148 | 0.084 |
| A3 | 1.122 | 0.134 | 0.081 | 1.126 | 0.138 | 0.084 | 1.124 | 0.141 | 0.085 |
| A4 | 1.100 | 0.113 | 0.068 | 1.126 | 0.130 | 0.081 | 1.075 | 0.095 | 0.054 |
| A5 | 1.132 | 0.145 | 0.088 | 1.141 | 0.155 | 0.094 | 1.072 | 0.093 | 0.053 |
| A6 | 1.133 | 0.126 | 0.080 | 1.126 | 0.133 | 0.082 | 1.094 | 0.104 | 0.063 |
| A7 | 1.143 | 0.141 | 0.089 | 1.145 | 0.158 | 0.096 | 1.098 | 0.117 | 0.068 |
| A8 | 1.196 | 0.214 | 0.129 | 1.130 | 0.144 | 0.086 | 1.078 | 0.100 | 0.057 |
| A9 | 1.140 | 0.151 | 0.092 | 1.127 | 0.147 | 0.087 | 1.101 | 0.121 | 0.070 |
| A10 | 1.134 | 0.136 | 0.086 | 1.126 | 0.132 | 0.083 | 1.100 | 0.110 | 0.068 |
| A11 | 1.162 | 0.162 | 0.101 | 1.130 | 0.144 | 0.087 | 1.083 | 0.103 | 0.059 |
| Mean | 1.134 | 0.141 | 0.087 | 1.133 | 0.145 | 0.088 | 1.093 | 0.112 | 0.065 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avramidou, E.V.; Korakaki, E.; Malliarou, E.; Boutsios, S. Studying the Genetic and the Epigenetic Diversity of the Endangered Species Juniperus drupacea Labill. towards Safeguarding Its Conservation in Greece. Forests 2023, 14, 1271. https://doi.org/10.3390/f14061271
Avramidou EV, Korakaki E, Malliarou E, Boutsios S. Studying the Genetic and the Epigenetic Diversity of the Endangered Species Juniperus drupacea Labill. towards Safeguarding Its Conservation in Greece. Forests. 2023; 14(6):1271. https://doi.org/10.3390/f14061271
Chicago/Turabian StyleAvramidou, Evangelia V., Evangelia Korakaki, Ermioni Malliarou, and Stefanos Boutsios. 2023. "Studying the Genetic and the Epigenetic Diversity of the Endangered Species Juniperus drupacea Labill. towards Safeguarding Its Conservation in Greece" Forests 14, no. 6: 1271. https://doi.org/10.3390/f14061271
APA StyleAvramidou, E. V., Korakaki, E., Malliarou, E., & Boutsios, S. (2023). Studying the Genetic and the Epigenetic Diversity of the Endangered Species Juniperus drupacea Labill. towards Safeguarding Its Conservation in Greece. Forests, 14(6), 1271. https://doi.org/10.3390/f14061271









