Genome-Wide Identification and Expression Profiling of B3 Transcription Factor Genes in Prunus armeniaca
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Analysis of B3 Superfamily Members in Apricot
2.2. Multiple Sequence Alignments and Phylogenetic Analysis
2.3. Gene Structure, Conserved Motif, and Domain Analysis
2.4. Chromosomal Locations and Synteny Analysis
2.5. Cis-Acting Elements and Gene Ontology (GO) Analysis
2.6. Expression Analysis Using RNA-Seq Data and Verification Using RT-qPCR
2.7. Oil Content Detection
3. Results
3.1. Identification of B3 Superfamily Genes in Apricot
3.2. Phylogenetic Analysis 3.2 Phylogenetic of Apricot B3 Genes
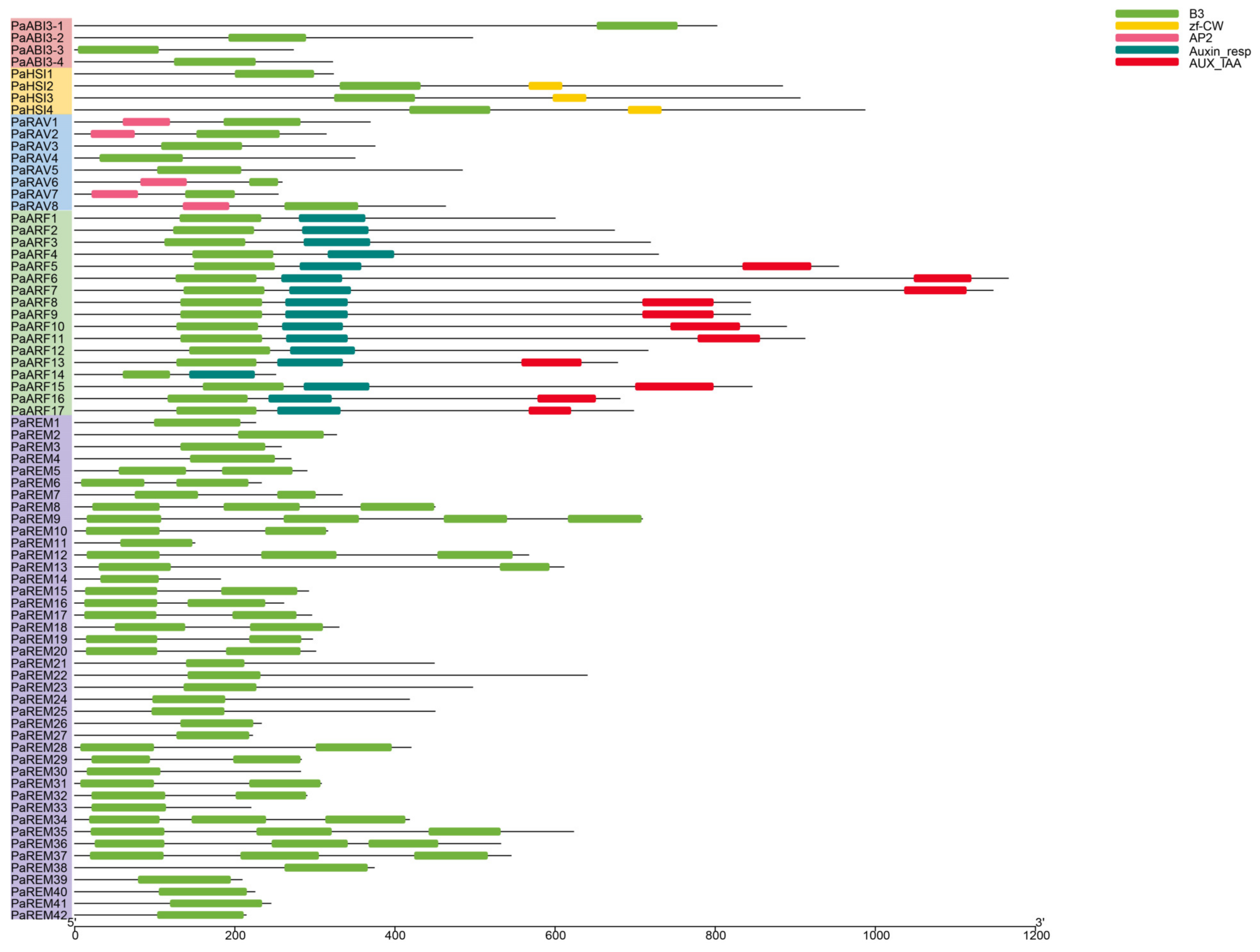
3.3. Gene Structure, Conserved Motif, and Domain Analysis of Apricot B3 Genes
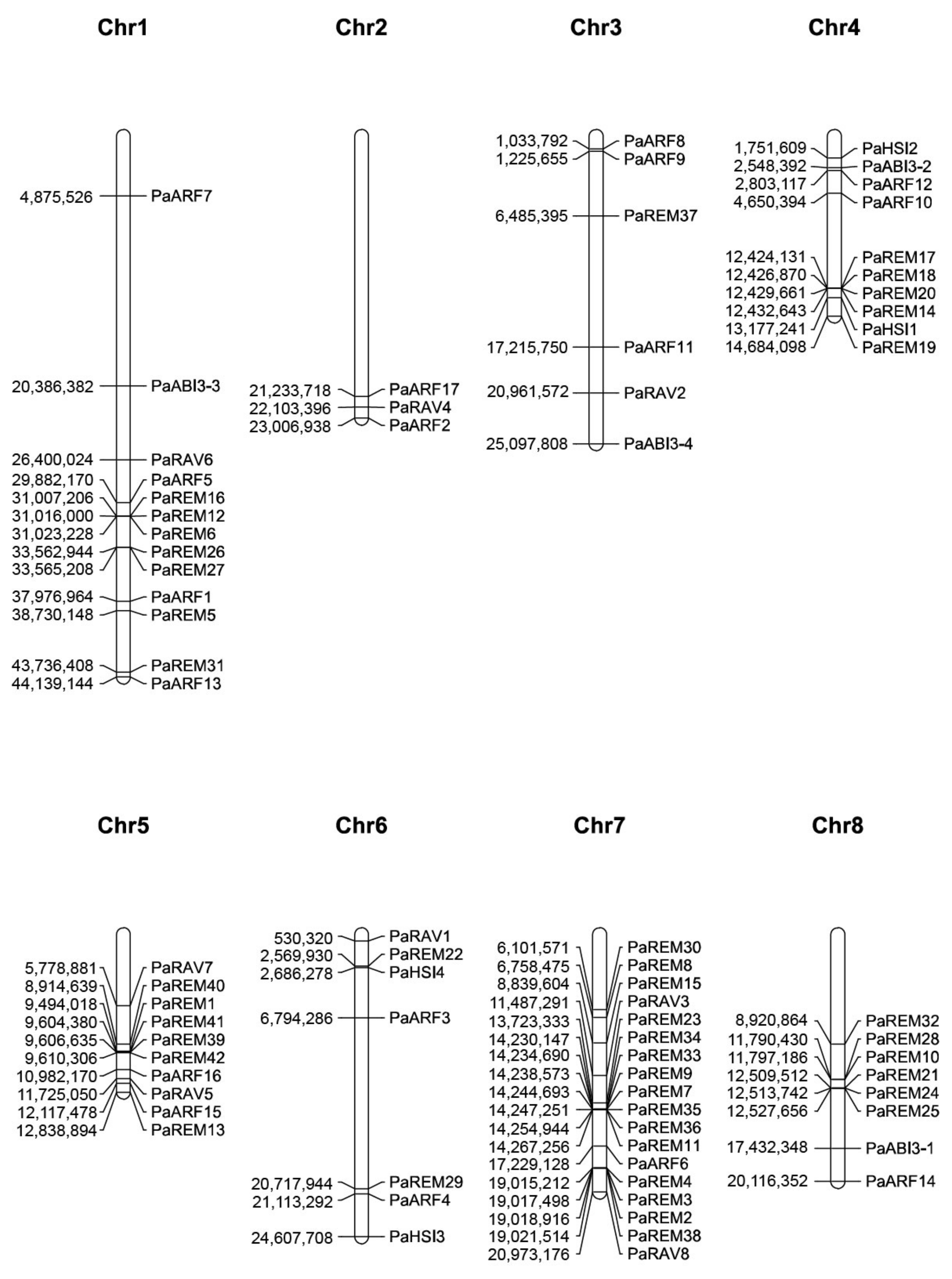
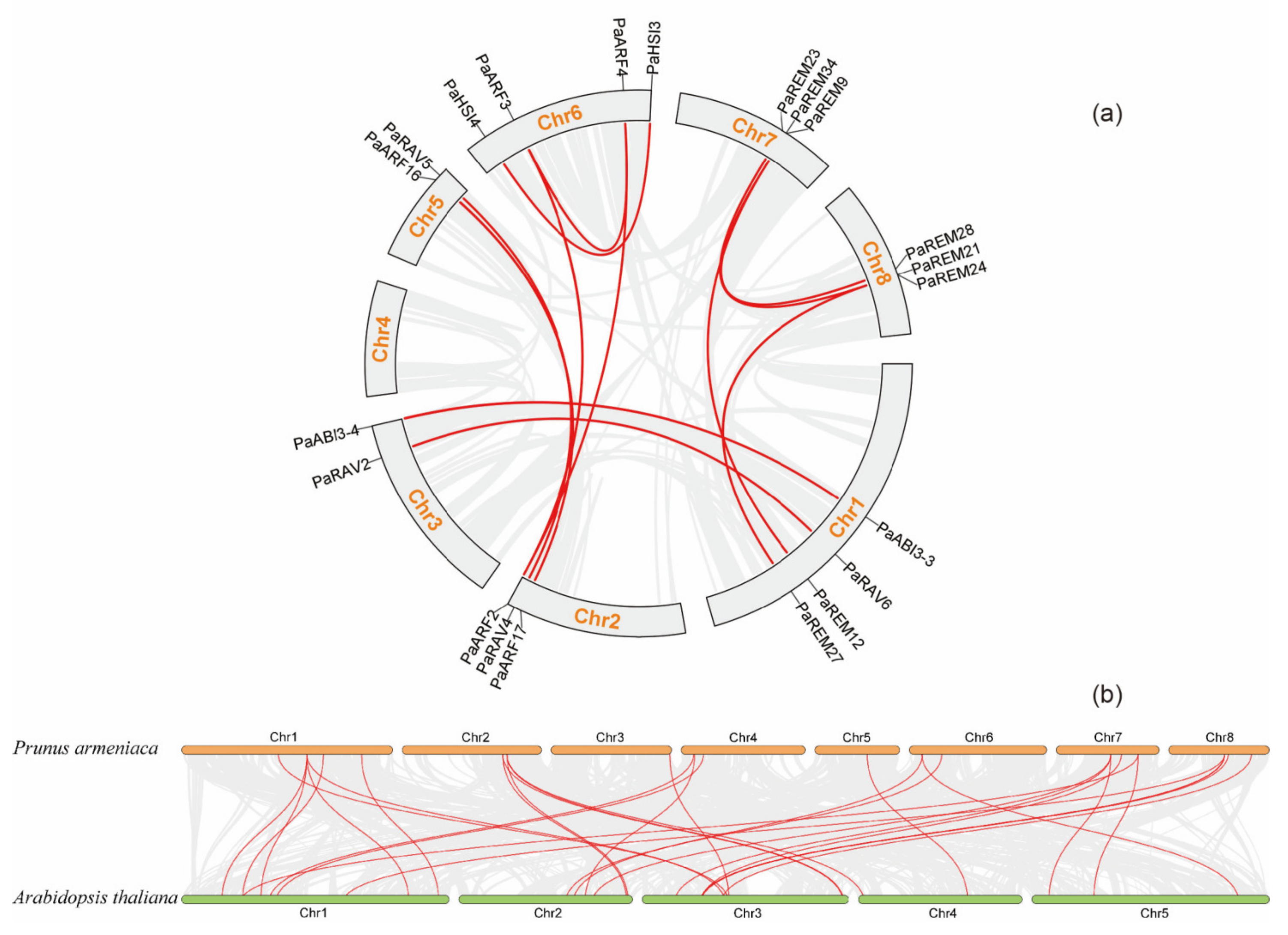
3.4. Chromosomal Locations and Synteny Analysis of Apricot B3 Genes
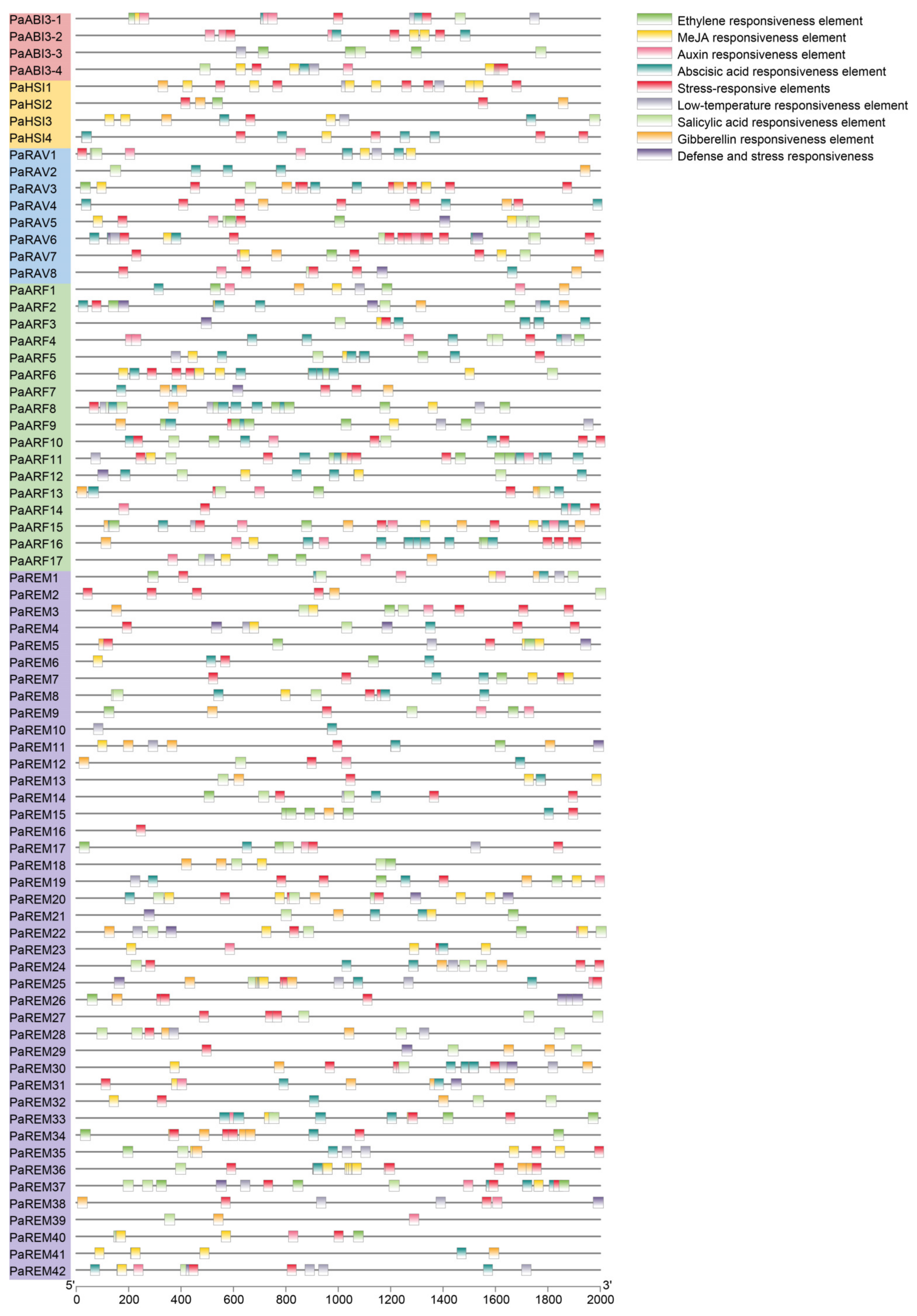
3.5. Cis-Acting Elements and Gene Ontology (GO) Analysis of Apricot B3 Genes
3.6. Tissue-Specific Expression Profiles and Identification of B3 Genes Associated with Oil Accumulation in Apricot
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Erdogan-Orhan, I.; Kartal, M. Insights into research on phytochemistry and biological activities of Prunus armeniaca L. (apricot). Food Res. Int. 2011, 44, 1238–1243. [Google Scholar] [CrossRef]
- Pawar, K.R.; Nema, P.K. Apricot kernel characterization, oil extraction, and its utilization: A review. Food Sci. Biotechnol. 2023, 32, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, K.; Kigawa, T.; Seki, M.; Shinozaki, K.; Yokoyama, S. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar] [CrossRef]
- Bianchi, V.J.; Manuel, R.; Livio, T.; Ignazio, V.; Claudio, B.; Pedro, M.N.G. Prunus transcription factors: Breeding perspectives. Front. Plant Sci. 2015, 6, 443. [Google Scholar] [CrossRef] [Green Version]
- Sasnauskas, G.; Manakova, E.; Lapėnas, K.; Kauneckaitė, K.; Siksnys, V. DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. FEBS J. 2018, 285, 4041–4059. [Google Scholar] [CrossRef] [Green Version]
- McCarty, D.R.; Hattori, T.; Carson, C.B.; Vasil, V.; Lazar, M.; Vasil, I.K. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 2018, 66, 895–905. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Zhang, R.; Wang, S.; Bian, Y.; Yin, Z. Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Mol. Biol. Rep. 2012, 39, 6267–6282. [Google Scholar] [CrossRef]
- Yamasaki, K. Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. Plant Cell 2012, 16, 3448–3459. [Google Scholar] [CrossRef] [Green Version]
- Peng, F.Y.; Weselake, R.J. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet. 2013, 126, 1305–1319. [Google Scholar] [CrossRef]
- Verma, S.; Bhatia, S. A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum L.). 3 Biotech 2019, 9, 346. [Google Scholar] [CrossRef]
- Chen, C.; Li, Y.; Zhang, H.; Ma, Q.; Wei, Z.; Chen, J.; Sun, Z. Genome-wide analysis of the RAV transcription factor genes in rice reveals their response patterns to hormones and virus infection. Viruses 2021, 13, 752. [Google Scholar] [CrossRef]
- Liu, Z.; Ge, X.X.; Wu, X.M.; Xu, Q.; Guo, W.W. Genome-wide analysis of the citrus B3 superfamily and their association with somatic embryogenesis. BMC Genom. 2020, 21, 305. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell Online 2019, 9, 799–807. [Google Scholar]
- Tsukagoshi, H.; Saijo, T.; Shibata, D.; Nakamura, M.K. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 2019, 138, 675–685. [Google Scholar] [CrossRef] [Green Version]
- Ulmasov, T.; Murfett, J.; Guilfoyle, H. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 11, 1963–1971. [Google Scholar]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Cubas, P.; Jarillo, J.A.; Fernandez-Calvin, B.; Salinas, J.; Martinez-Zapater, J.M. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiol. 2002, 128, 418–427. [Google Scholar] [CrossRef]
- Bies-Etheve, N.; da Silva Conceicao, A.; Giraudat, J.; Koornneef, M.; Leon-Kloosterziel, K.; Valon, C.; Delseny, M. Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol. Biol. 1999, 40, 1045–1054. [Google Scholar] [CrossRef]
- Swaminathan, K.; Peterson, K.; Jack, T. The plant B3 superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef]
- Suzuki, M.; Wang, H.Y.; McCarty, D.R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 Genes. Plant Physiol. 2006, 143, 902–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999, 19, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Guilfoyle, H.T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Magnani, E.; Sjölander, K.; Hake, S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef] [Green Version]
- Mantegazza, O.; Gregis, V.; Mendes, M.A.; Morandini, P.; Alves-Ferreira, M.; Patreze, C.M.; Nardeli, S.M.; Kater, M.M.; Colombo, L. Analysis of the Arabidopsis REM gene family predicts functions during flower development. Ann. Bot. 2014, 114, 1507–1515. [Google Scholar] [CrossRef] [Green Version]
- Monke, G.; Altschmied, L.; Tewes, A.; Reidt, W.; Mock, H.P.; Baumlein, H.; Conrad, U. Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 2004, 219, 158–166. [Google Scholar] [CrossRef]
- Parcy, F.; Valon, C.; Kohara, A.; Misera, S.; Giraudat, J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell Online 1997, 9, 1265–1277. [Google Scholar]
- Crowe, A.J.; Abenes, M.; Plant, A.; Moloney, M.M. The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 2000, 151, 171–181. [Google Scholar] [CrossRef]
- Grimault, A.; Gendrot, G.; Chaignon, S.; Gilard, F.; Tcherkez, G.; Thevenin, J.; Dubreucq, B.; Depege-Fargeix, N.; Rogowsky, P.M. Role of B3 domain transcription factors of the AFL family in maize kernel filling. Plant Sci. 2015, 236, 116–125. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Bedi, S.; Nag Chaudhuri, R. Transcription factor ABI3 auto-activates its own expression during dehydration stress response. FEBS Lett. 2018, 592, 2594–2611. [Google Scholar] [CrossRef] [Green Version]
- Tamminen, I.; Makela, P.; Heino, P.; Palva, E.T. Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana. Plant J. 2001, 25, 1–8. [Google Scholar]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N.; et al. ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef] [Green Version]
- Chhun, T.; Chong, S.Y.; Park, B.S.; Wong, E.C.C.; Yin, J.L.; Kim, M.; Chua, N.H. HSI2 repressor recruits MED13 and HDA6 to down-regulate seed maturation gene expression directly during Arabidopsis early seedling growth. Plant Cell Physiol. 2016, 57, 1689–1706. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Wang, H.; Abdelmageed, H.; Veerappan, V.; Tadege, M.; Allen, R.D. HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 2020, 227, 840–856. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.G.; Dinh, T.T.; Li, D.M.; Shi, B.H.; Li, Y.P.; Cao, X.W.; Guo, L.; Pan, Y.Y.; Jiao, Y.L.; Chen, X.M. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014, 80, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Muto, H.; Higuchi, K.; Matamura, T.; Tatematsu, K.; Koshiba, T.; Yamamoto, K.T. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004, 40, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Li, K.X.; Chen, L.; Zou, Y.M.; Liu, H.P.; Tian, Y.P.; Li, D.X.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. Microrna167-directed regulation of the auxin response factors, GmARF8a and GmARF8b, is required for soybean (Glycine max L.) nodulation and lateral root development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 2011, 285, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Khurana, J.P. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009, 276, 3148–3162. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Sun, T.T.; Yang, S.J.; Wang, X.; Chao, J.T.; Li, X.X.; Hu, J.H.; Cui, M.M.; Liu, G.S.; Wang, D.W.; et al. Insight into the B3 transcription factor superfamily and expression profiling of B3 genes in axillary buds after topping in tobacco (Nicotiana tabacum L.). Genes 2019, 10, 164. [Google Scholar] [CrossRef] [Green Version]
- Osnato, M.; Castillejo, C.; Matias-Hernandez, L.; Pelaz, S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. Plant Cell 2015, 27, 620–632. [Google Scholar] [CrossRef] [Green Version]
- Sohn, K.; Lee, S.; Jung, H.; Hong, J.; Hwang, B. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006, 61, 897–915. [Google Scholar] [CrossRef]
- Levy, Y.Y. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef]
- Caselli, F.; Beretta, V.M.; Mantegazza, O.; Petrella, R.; Leo, G.; Guazzotti, A.; Herrera-Ubaldo, H.; de Folter, S.; Mendes, M.A.; Kater, M.M.; et al. REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1351. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Y.; Wu, S.L.; Sun, J.J.; Sun, J.R.; Wang, H.L.; Cao, X.; Lu, J.; Jalal, A.; Wang, C.Q. Genome-wide analysis reveals widespread roles for RcREM genes in floral organ development in Rosa chinensis. Genomics 2021, 113, 3881–3894. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef] [PubMed]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids. Res. 2012, 40, 49. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.P.; Lin, J.J.; Li, W.H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 2016, 6, 25164. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Nvsvrot, T.; Wang, N. Comparative transcriptomic analysis uncovers conserved pathways involved in adventitious root formation in poplar. Physiol. Mol. Biol. Plants 2021, 27, 1903–1918. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lu, Y.; Wang, J.; Hu, J.; Wuyun, T. Genome-wide screening of long non-coding RNAs involved in rubber biosynthesis in Eucommia ulmoides. Integr. Plant Biol. 2018, 60, 1070–1082. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Cui, B.; Zhu, H.; Phommakoun, B.; Zhang, D.; Li, Y.M.; Zhao, F.; Zhao, Z. Accumulation pattern of amygdalin and prunasin and its correlation with fruit and kernel agronomic characteristics during apricot (Prunus armeniaca L.) kernel development. Foods 2021, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.C.; Chen, Z.; Hu, F.C.; Fan, W.; Wang, X.H.; Guo, L.J.; Fan, H.Y.; Luo, Z.W.; Zhang, Z.L. Genome-wide characterization and expression profiling of B3 superfamily during ethylene-induced flowering in pineapple (Ananas comosus L.). BMC Genom. 2021, 22, 561. [Google Scholar] [CrossRef]
- Song, L.; Wu, S.; Tsang, A. Phylogenetic Analysis of Protein Family. Methods Mol. Biol. (Clifton N. J.) 2018, 1775, 267–275. [Google Scholar]
- Wang, Y.; You, F.M.; Lazo, G.R.; Luo, M.C.; Thilmony, R.; Gordon, S.; Kianian, S.F.; Gu, Y.Q. PIECE: A database for plant gene structure comparison and evolution. Nucleic Acids Res. 2013, 41, 1159–1166. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.B.; Ao, T.; Zhang, Y.Y.; Wu, D.; Xu, W.; Han, B.; Liu, A.Z. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis). Plant Divers. 2022, 44, 201–212. [Google Scholar] [CrossRef]
- Ahmad, B. Genomic Organization of the B3-Domain Transcription factor family in grapevine (Vitis vinifera L.) and expression during seed development in seedless and seeded cultivars. Int. J. Mol. Sci. 2019, 20, 4553. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Rensing, S.A. Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 2014, 17, 43–48. [Google Scholar] [CrossRef]
- Romanel, E.A.C.; Schrago, C.G.; Counago, R.M.; Russo, C.A.M.; Alves-Ferreira, M. Evolution of the B3 DNA binding superfamily: New insights into REM family gene diversification. PLoS ONE 2009, 4, 5791. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Jiwan, D.; Cho, I.; Lee, T.; Abbott, A.; Sosinski, B.; Main, D. Synteny of Prunus and other model plant species. BMC Genom. 2009, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.W.; Wang, Y.F.; Zhang, S.M.; Xie, K.L.; Zhang, C.; Xi, Y.J.; Sun, F.L. Genome-wide analysis of the abiotic stress-related bZIP family in switchgrass. Mol. Biol. Rep. 2020, 47, 4439–4454. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J.A. Genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Salih, H.; Tan, L.; Htet, N.N.W. Genome-Wide identification, characterization of bHLH transcription factors in mango. Trop. Plant Biol. 2021, 14, 72–81. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Brady, S.M.; Sarkar, S.F.; Bonetta, D.; Mccourt, P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003, 34, 67–75. [Google Scholar] [CrossRef]
- Liu, X.D.; Zhang, H.; Zhao, Y.; Feng, Z.Y.; Li, Q.; Yang, H.Q.; Luan, S.; Li, J.M.; He, Z.H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Wang, L.; Zhang, Y.X.; Zhu, G.P.; Zhu, X.C.; Xia, Y.X.; Li, J.B.; Gao, X.; Wang, S.L.; Zhang, J.H.; et al. Genome-wide identification and function of aquaporin genes during dormancy and sprouting periods of kernel-using apricot (Prunus armeniaca L.). Front. Plant Sci. 2021, 12, 690040. [Google Scholar] [CrossRef] [PubMed]
- Azuaje, F.; Al-Shahrour, F.; Dopazo, J. Ontology-driven approaches to analyzing data in functional genomics. Bioinform. Drug Discov. 2006, 316, 67–86. [Google Scholar] [CrossRef]
- Zhou, H.B.; Song, Z.H.; Zhong, S.; Zuo, L.Y.; Qi, Z.; Qu, L.J.; Lai, L.H. Mechanism of DNA-induced phase separation for transcriptional repressor VRN1. Angew. Chem. 2019, 131, 4912–4916. [Google Scholar] [CrossRef]
- Sasnauskas, G.; Kauneckaitė, K.; Siksnys, V. Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1. Nucleic Acids Res. 2018, 46, 4316–4324. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.C.; Qiao, L.F.; Chen, J.C.; Rong, Y.H.; Zhao, Y.H.; Cui, X.K.; Xu, J.P.; Hou, X.M.; Dong, C.H. Arabidopsis REM16 acts as a B3 domain transcription factor to promote flowering time via directly binding to the promoters of SOC1 and FT. Plant J. 2020, 103, 1386–1398. [Google Scholar] [CrossRef]
- Revalska, M.; Vassileva, V.; Zehirov, G.; Goormachtig, S.; Iantcheva, A. Assessment of the function and expression pattern of auxin response factor B3 in the model legume plant Medicago truncatula. Turk. J. Biol. 2017, 41, 66–76. [Google Scholar] [CrossRef]
- Shao, J.; Liu, X.; Wang, R.; Zhang, G.; Yu, F. The over-expression of an Arabidopsis B3 transcription factor, ABS2/NGAL1, leads to the loss of flower petals. PLoS ONE 2012, 7, 49861. [Google Scholar] [CrossRef] [Green Version]
- Tsukagoshi, H.; Morikami, A.; Nakamura, K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 2007, 104, 2543–2547. [Google Scholar] [CrossRef]
- Matias-Hernandez, L.; Aguilar-Jaramillo, A.E.; Marin-Gonzalez, E.; Suarez-Lopez, P.; Pelaz, S. RAV genes: Regulation of floral induction and beyond. Ann. Bot. 2014, 114, 1459–1470. [Google Scholar] [CrossRef]
- Hu, Y.X.; Wang, Y.X.; Liu, X.F.; Li, J.Y. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004, 14, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Woo, H.R.; Kim, J.H.; Kim, J.; Kim, J.; Lee, U.; Song, I.J.; Kim, J.H.; Lee, H.Y.; Nam, H.G.; Lim, P.O. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 2010, 61, 3947–3957. [Google Scholar] [CrossRef] [Green Version]
- Turan, S.; Topcu, A.; Karabulut, I.; Vural, H.; Hayaloglu, A.A. Fatty acid, triacylglycerol, phytosterol, and tocopherol variations in kernel oil of Malatya apricots from Turkey. J. Agric. Food Chem. 2007, 55, 10787–10794. [Google Scholar] [CrossRef]
- Popa, V.M.; Bele, C.; Poiana, M.A.; Dumbrava, D.; Jianu, C. Evaluation of bioactive compounds and of antioxidant properties in some oils obtained from food industry by-products. Rom. Biotechnol. Lett. 2011, 16, 6234–6241. [Google Scholar]
- Aziz, U.; Tang, T.; Saleem, N.; Yang, Z.; Zhang, M. New insights revealed the evolution of the AFL subfamily of B3 transcription factors from chlorophyta and its requisite in land plants. Pak. J. Agric. Sci. 2020, 57, 1219–1235. [Google Scholar]
- Yang, Z.; Liu, X.L.; Wang, K.; Li, Z.W.; Jia, Q.L.; Zhao, C.Z.; Zhang, M. ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 2022, 73, 2077–2092. [Google Scholar] [CrossRef]
- Mönke, G.; Seifert, M.; Keilwagen, J.; Mohr, M.; Grosse, I.; Hähnel, U.; Junker, A.; Weisshaar, B.; Conrad, U.; Bäumlein, H.; et al. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 2012, 40, 8240–8254. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Schmidt, M.A.; Collet, K.H.; Liu, Z.B.; Coy, M.; Abbitt, S.; Molloy, L.; Frank, M.; Everard, J.D.; Booth, R.; et al. RNAi and CRISPR-Cas silencing E3-RING ubiquitin ligase AIP2 enhances soybean seed protein content. J. Exp. Bot. 2022, 73, 7285–7297. [Google Scholar] [CrossRef]
- Singer, S.D.; Jayawardhane, K.N.; Jiao, C.; Weselake, R.J.; Chen, G. The effect of AINTEGUMENTA-LIKE 7 over-expression on seed fatty acid biosynthesis, storage oil accumulation and the transcriptome in Arabidopsis thaliana. Plant Cell Rep. 2021, 40, 1647–1663. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Yu, W.; Wang, L.; Zhao, H.; Hu, J.; Wuyun, T.; Liu, H. Genome-Wide Identification and Expression Profiling of B3 Transcription Factor Genes in Prunus armeniaca. Forests 2023, 14, 1523. https://doi.org/10.3390/f14081523
Shi X, Yu W, Wang L, Zhao H, Hu J, Wuyun T, Liu H. Genome-Wide Identification and Expression Profiling of B3 Transcription Factor Genes in Prunus armeniaca. Forests. 2023; 14(8):1523. https://doi.org/10.3390/f14081523
Chicago/Turabian StyleShi, Xiaodan, Wanwen Yu, Lin Wang, Han Zhao, Jingjing Hu, Tana Wuyun, and Huimin Liu. 2023. "Genome-Wide Identification and Expression Profiling of B3 Transcription Factor Genes in Prunus armeniaca" Forests 14, no. 8: 1523. https://doi.org/10.3390/f14081523
APA StyleShi, X., Yu, W., Wang, L., Zhao, H., Hu, J., Wuyun, T., & Liu, H. (2023). Genome-Wide Identification and Expression Profiling of B3 Transcription Factor Genes in Prunus armeniaca. Forests, 14(8), 1523. https://doi.org/10.3390/f14081523






