Phenotypic Differences of Leaves and Transcriptome Analysis of Fraxinus mandshurica × Fraxinus sogdiana F1 Variety
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Determination of the Leaf Phenotype
2.3. Determination of Photosynthetic Parameters of Leaves
2.4. Complementary DNA Library Construction and RNA Sequencing
2.5. Weighted Gene Coexpression Network Analysis (WCGNA) for Leaf-Specific Expressed Genes
2.6. Validation of RNA-Seq Data by Quantitative Real-Time PCR (qRT-PCR)
2.7. Data Processing and Statistics
3. Results
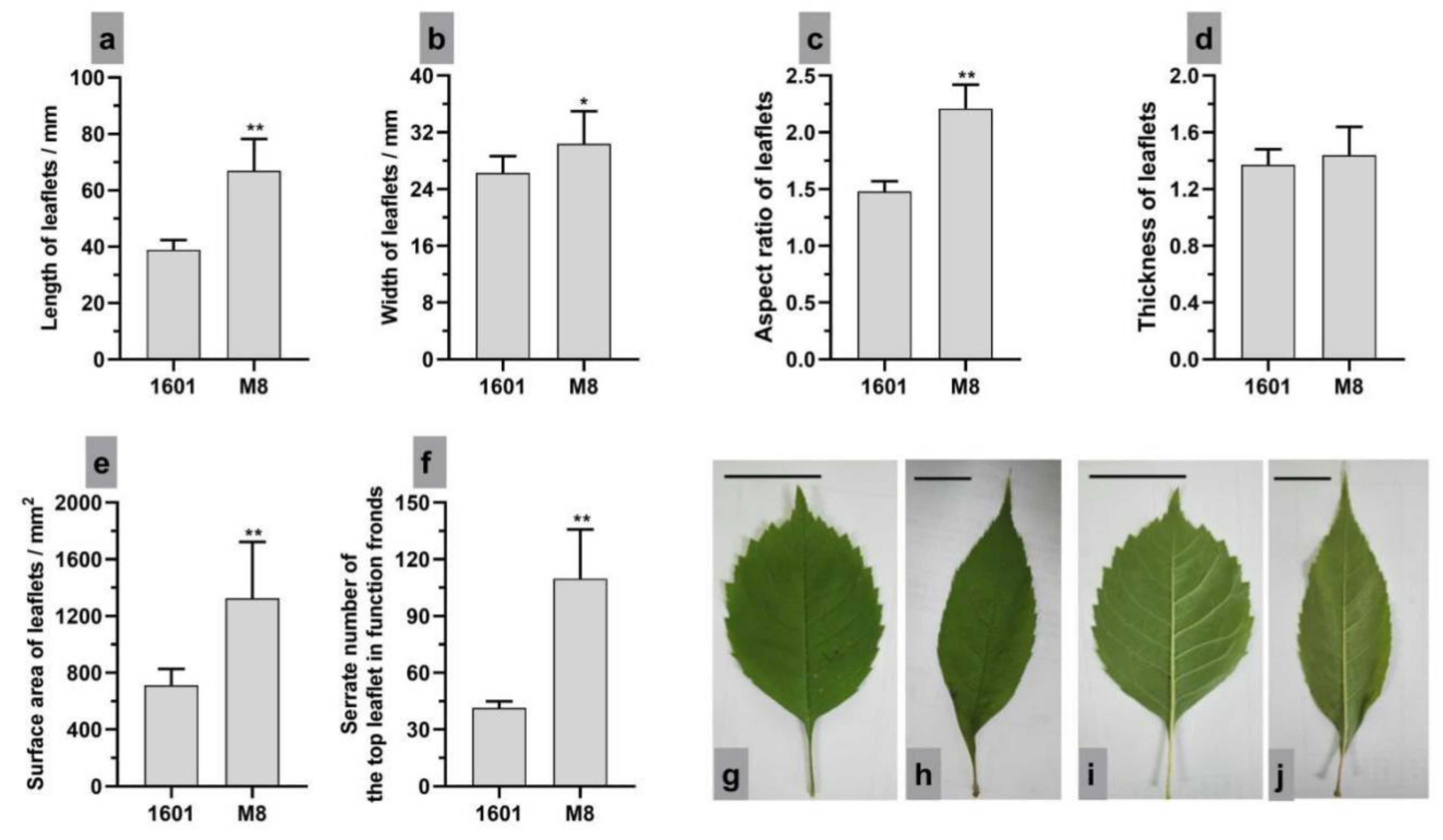
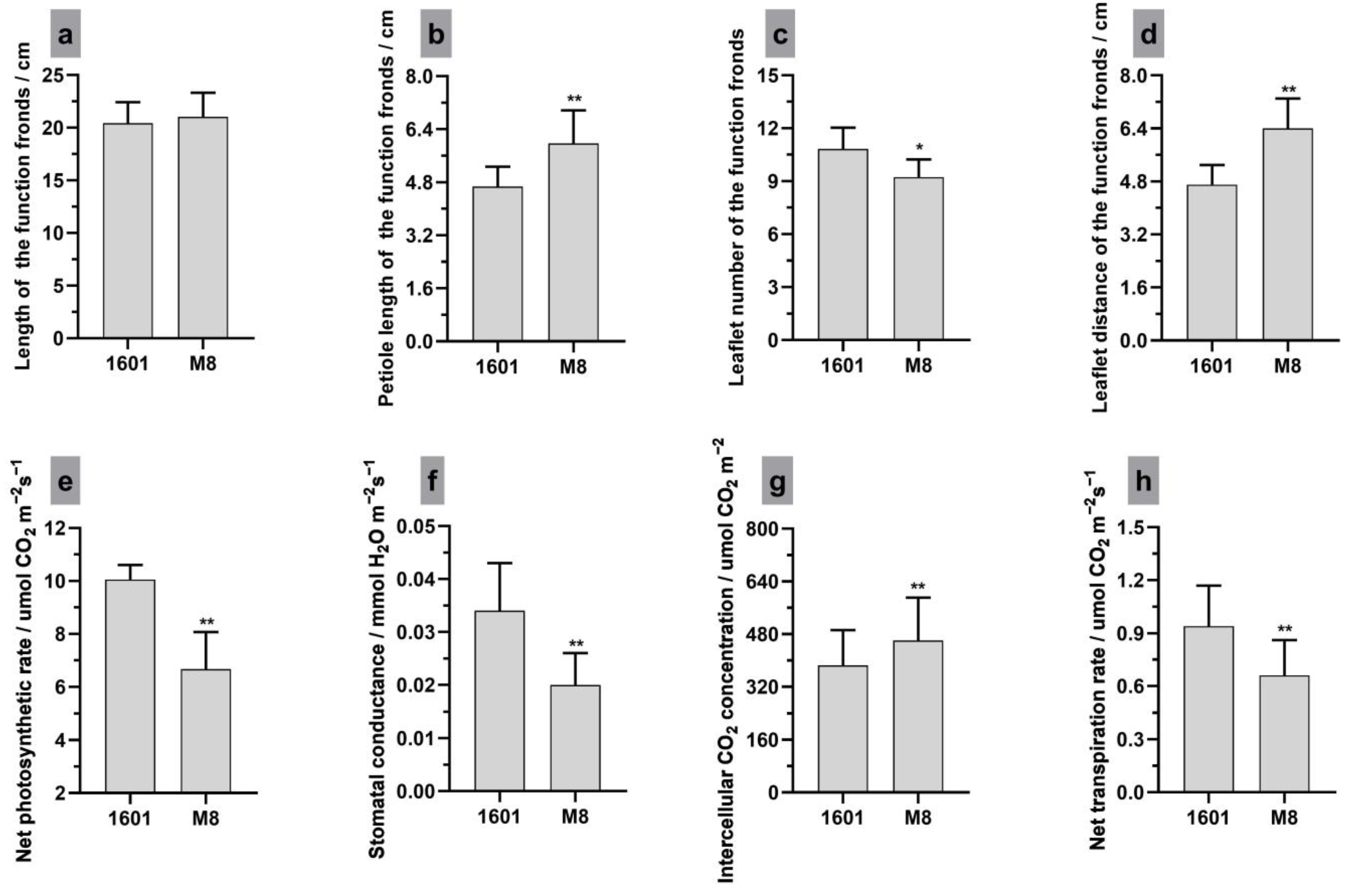
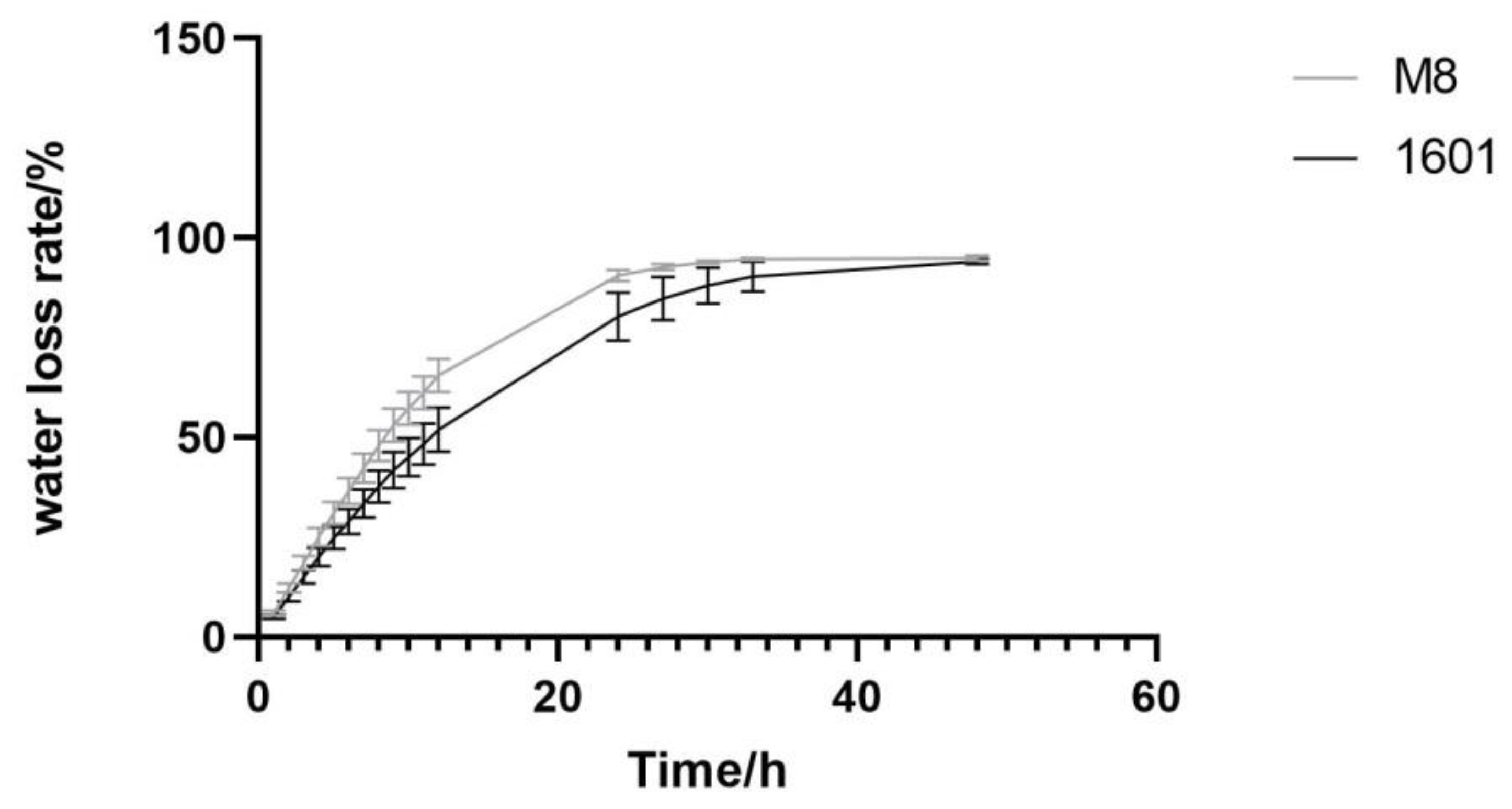
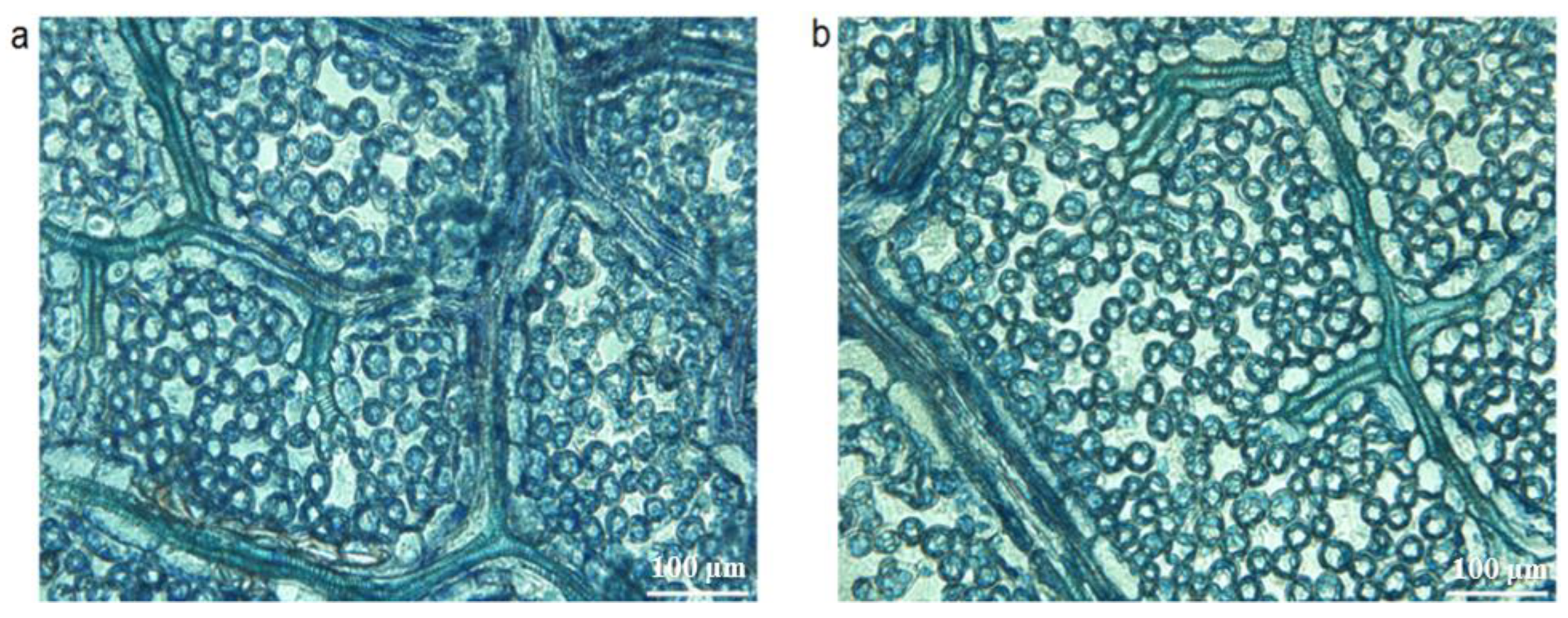
3.1. Leaf Morphology, Photosynthetic Parameters and Microstructure of Interspecific Hybrid (1601) and Its Female Parent (M8)
3.2. Transcriptome Sequencing of M8 and 1601 Leaves
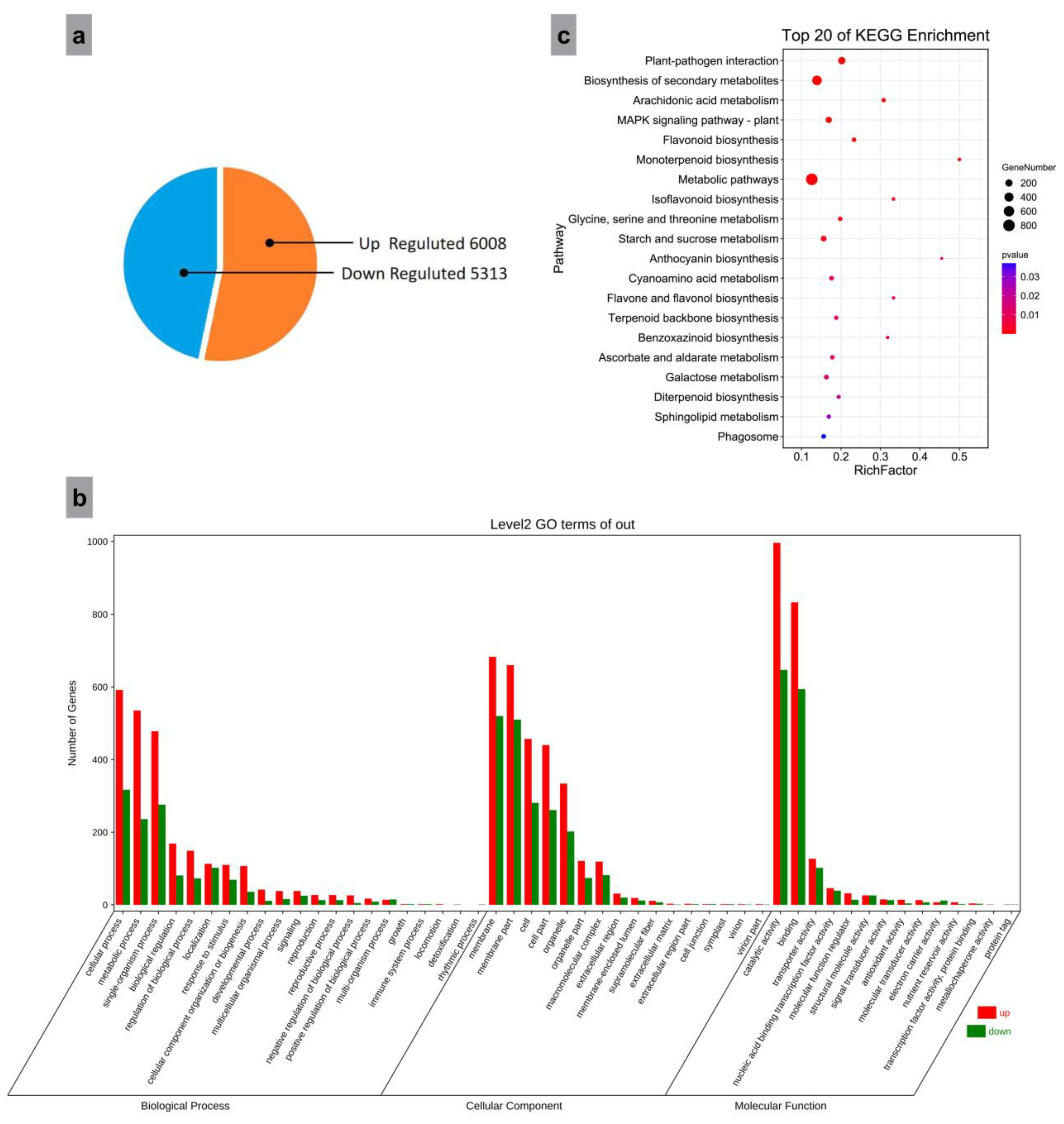
3.3. Screening for Differentially Expressed Genes (DEGs)
3.4. Gene Ontology (GO) Enrichment Analysis of DEGs
3.5. KEGG Pathway Enrichment Analysis of DEGs
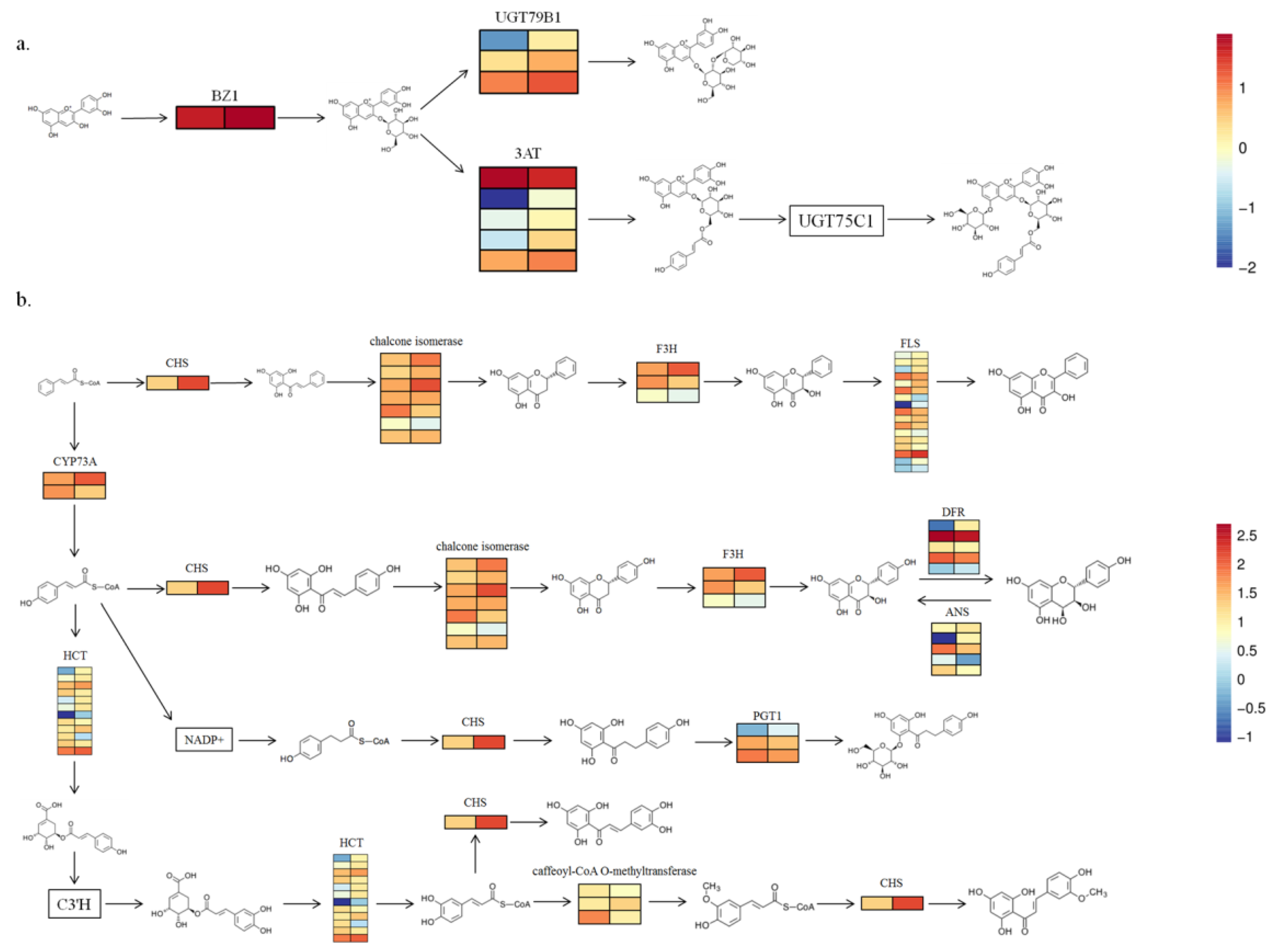
3.6. Biosynthesis Genes of Anthocyanins and Flavonoids Are Involved in the Development of Plant Leaves
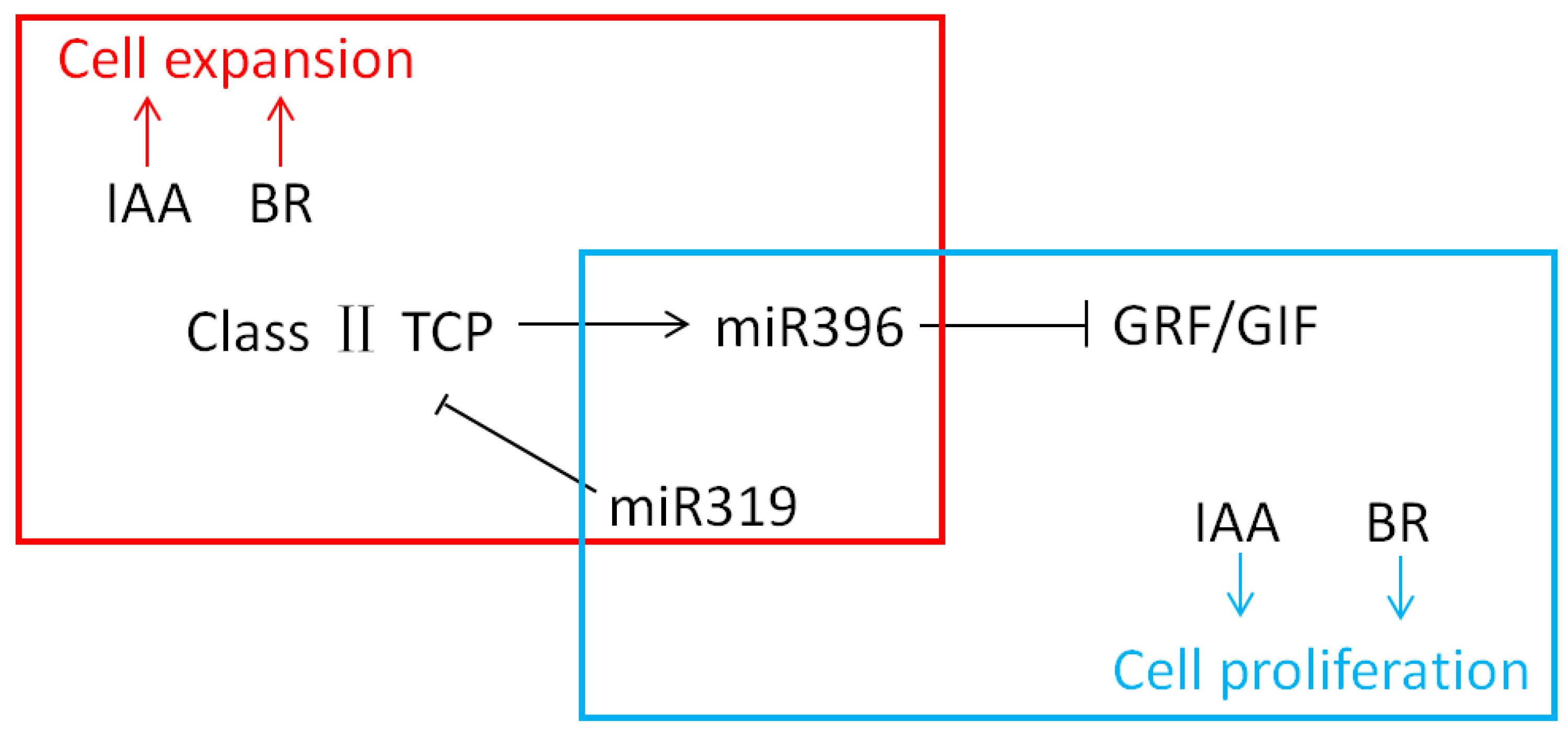
3.7. Auxin and Brassinolides (BRs) Are Involved in the Regulation of Plant-Leaf Development
3.8. Differences in Transcription Factors Associated with Leaf Development between 1601 and M8
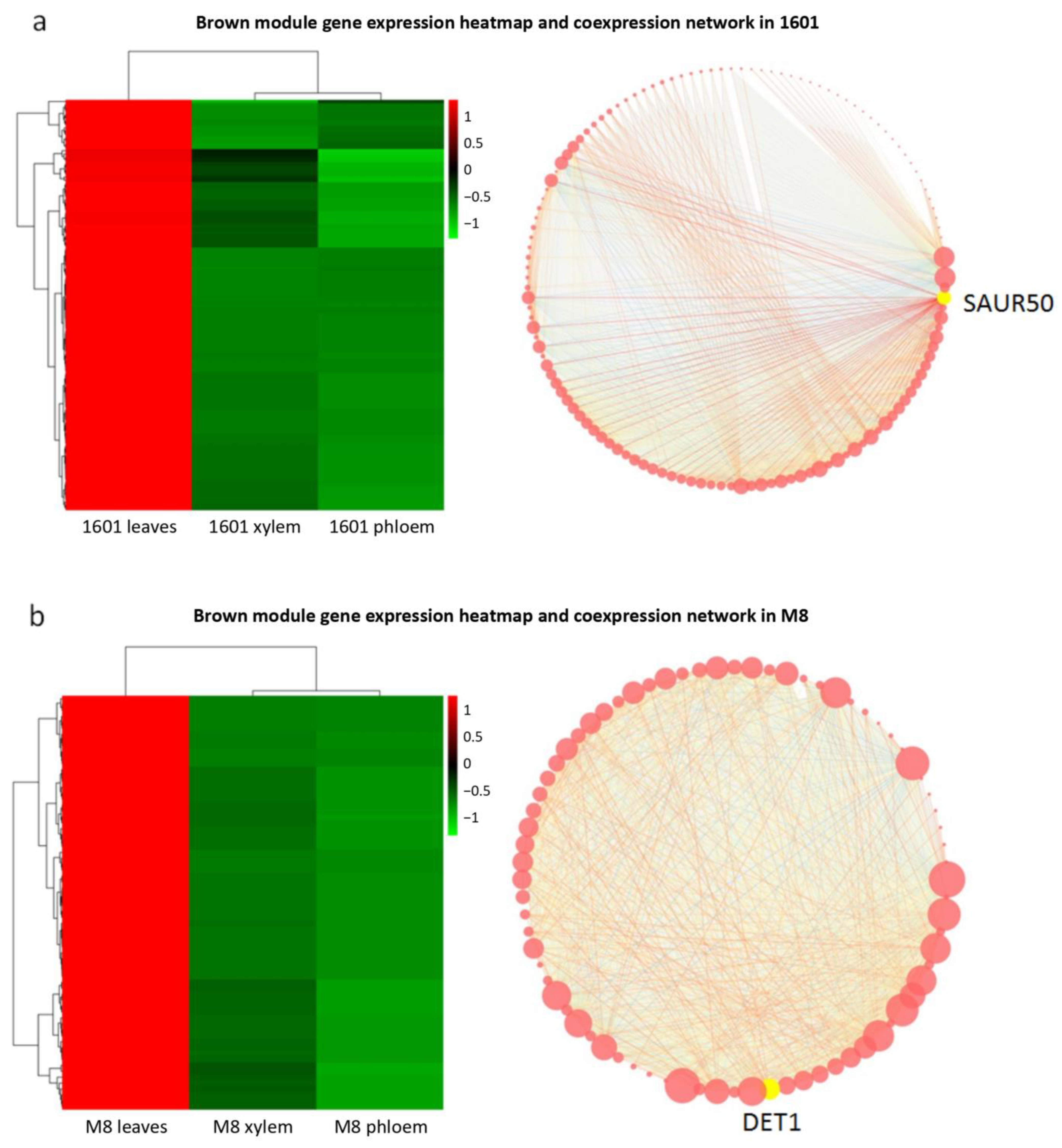
3.9. Association Analysis between Modules and Different Parts in Gene Co-Expression Network
3.10. Mining of Core Genes and Construction of Interaction Networks
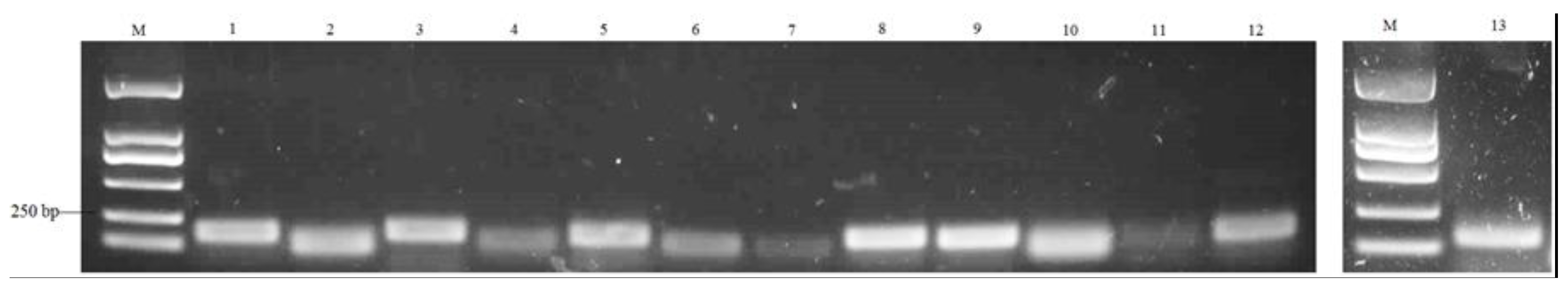
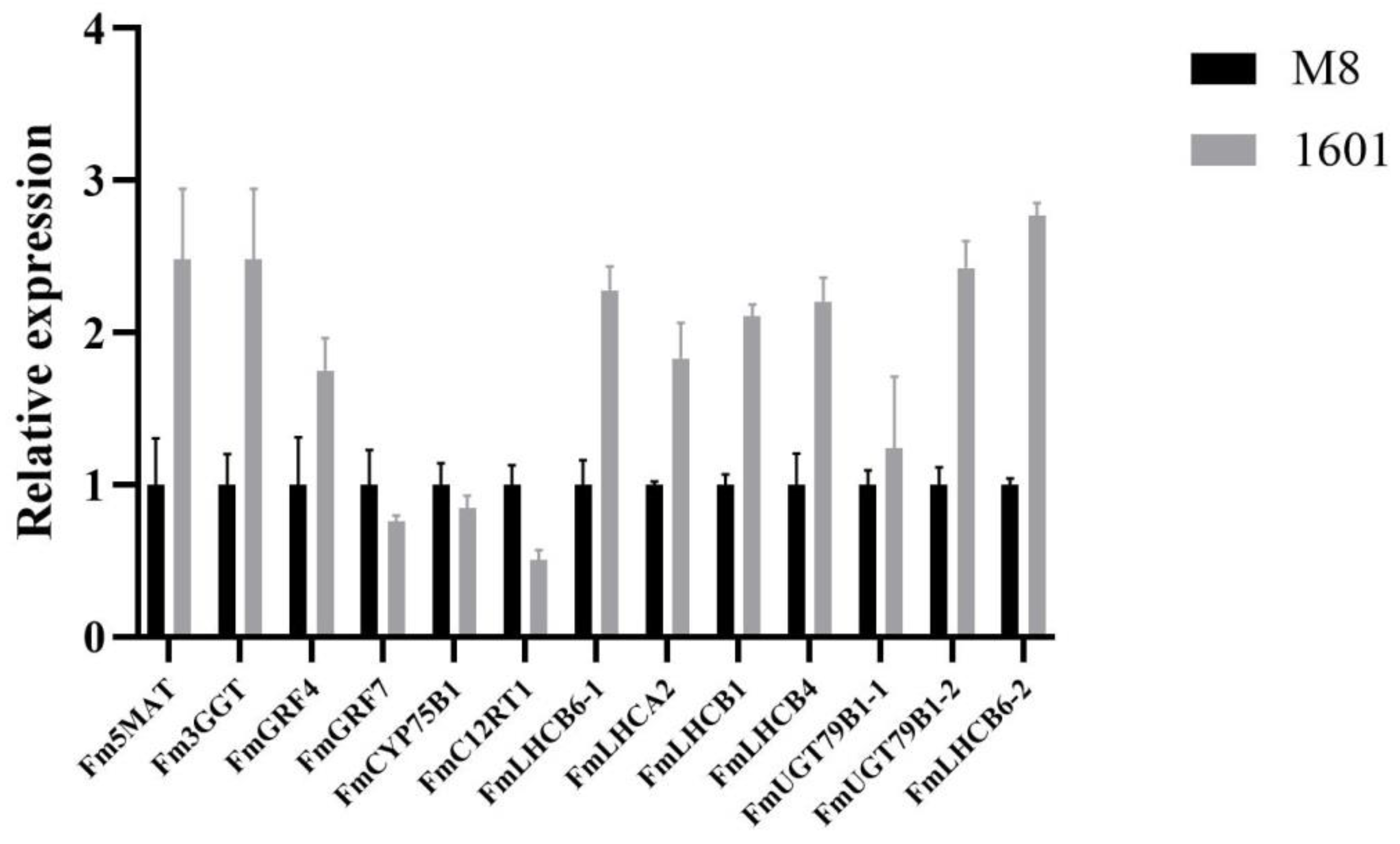
3.11. Quantitative Real-Time PCR (qRT-PCR) Validation of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nicotra, A.B.; Leigh, A.; Boyce, C.K.; Jones, C.S.; Niklas, K.J.; Royer, D.L.; Tsukaya, H. The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 2011, 38, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. Leaf shape: Genetic controls and environmental factors. Int. J. Dev. Biol. 2005, 49, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Kalve, S.; De Vos, D.; Beemster, G.T. Leaf development: A cellular perspective. Front. Plant Sci. 2014, 5, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarvepalli, K.; Das Gupta, M.; Challa, K.R.; Nath, U. Molecular cartography of leaf development—Role of transcription factors. Curr. Opin. Plant Biol. 2019, 47, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champagne, C.; Sinha, N. Compound leaves: Equal to the sum of their parts? Development 2004, 131, 4401–4412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warman, L.; Moles, A.T.; Edwards, W. Not so simple after all: Searching for ecological advantages of compound leaves. Oikos 2011, 120, 813–821. [Google Scholar] [CrossRef]
- Rubén, M. Compound leaves and the evolution of leaf size and display. Ideas Ecol. Evol. 2012, 5, 6–8. [Google Scholar] [CrossRef]
- Chen, R. Functional Genomics and Genetic Control of Compound Leaf Development in Medicago truncatula: An Overview. Methods Mol. Biol. 2018, 1822, 197–203. [Google Scholar] [CrossRef]
- Macfarlane, D.W.; Meyer, S.P. Characteristics and distribution of potential ash tree hosts for emerald ash borer—ScienceDirect. For. Ecol. Manag. 2005, 213, 15–24. [Google Scholar] [CrossRef]
- Li, B.; Howe, G.T.; Wu, R. Developmental factors responsible for heterosis in aspen hybrids (Populus tremuloides × P. tremula). Tree Physiol. 1998, 18, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eric, J.R.; Daniel, A.H.; David, R.S. Interspecific Variation in Resistance to Emerald Ash Borer (Coleoptera: Buprestidae) Among North American and Asian Ash (Fraxinus spp.). Environ. Entomol. 2008, 37, 242–246. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, M.; Kasem, S.; Lee, D.J.; Henry, R. Mapping species differences for adventitious rooting in a Corymbia torelliana × Corymbia citriodora subspecies variegata hybrid. Tree Genet. Genomes 2008, 4, 715–725. [Google Scholar] [CrossRef]
- Cheng, X.; Chen, S.; Chen, F.; Fang, W.; Deng, Y.; She, L. Interspecific hybrids between Dendranthema morifolium (Ramat.) Kitamura and D. nankingense (Nakai) Tzvel. achieved using ovary rescue and their cold tolerance characteristics. Euphytica 2010, 172, 101–108. [Google Scholar] [CrossRef]
- Wang, J.; Huang, L.; Bao, M.; Liu, G. Production of interspecific hybrids between Lilium longiflorum and L. lophophorum var. linearifolium via Ovule Culture at Early Stage. Euphytica 2009, 167, 45–55. [Google Scholar] [CrossRef]
- He, L.; Xu, Y.; Zeng, F.; Tian, H.; Xiao, Y.; Liu, H.; Yu, L.; Zhan, Y. Establishment of a micropropagation supporting technology for the Fraxinus mandshurica × Fraxinus sogdiana. Vitr. Cell. Dev. Biol.-Plant 2021, 57, 307–318. [Google Scholar] [CrossRef]
- He, Z.; Zhan, Y.; Zeng, F.; Zhao, X.T.; Wang, X. Drought physiology and gene expression characteristics of Fraxinus interspecific hybrids. Plant Growth Regul. 2016, 78, 179–193. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Thomasset, M.; Fernandez-Manjarres, J.F.; Douglas, G.C.; Frascaria-Lacoste, N.; Raquin, C.; Hodkinson, T.R. Molecular and morphological characterization of reciprocal F1 hybrid ash (Fraxinus excelsior × Fraxinus angustifoua, Oleaceae) and parental species reveals asymmetric character inheritance. Int. J. Plant Sci. 2011, 172, 423–433. [Google Scholar] [CrossRef]
- Lopez, J.L.; Abt, R.C.; Dvorak, W.S.; Hodge, G.R.; Phillips, R. Tree breeding model to assess financial performance of pine hybrids and pure species: Deterministic and stochastic approaches for South Africa. New For. 2018, 49, 123–142. [Google Scholar] [CrossRef]
- Zeng, F.; Zhou, S.; Zhan, Y.; Dong, J. Drought resistance and DNA methylation of interspecific hybrids between Fraxinus mandshurica and Fraxinus americana. Trees 2014, 28, 1679–1692. [Google Scholar] [CrossRef]
- Wu, S.; Xu, J.; Li, G.; Du, Z.; Lu, Z.; Li, B. Age trends and correlations of growth and wood properties in clone of Eucalyptus urophylla × E. grandis in Guangdong, China. J. For. Res. 2012, 23, 467–472. [Google Scholar] [CrossRef]
- Wang, R.; Gao, M.; Ji, S.; Wang, S.; Meng, Y.; Zhou, Z. Carbon allocation, osmotic adjustment, antioxidant capacity and growth in cotton under long-term soil drought during flowering and boll-forming period. Plant Physiol. Biochem. 2016, 107, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Sandquist, D.R.; Ehleringer, J.R. Population- and family-level variation of brittlebush (Encelia farinosa, Asteraceae) pubescence: Its relation to drought and implications for selection in variable environments. Am. J. Bot. 2003, 90, 1481–1486. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Kong, F.; Zhuo, C. From genes to networks: The genetic control of leaf development. J. Integr. Plant Biol. 2021, 63, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L.; Hohtola, A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010, 33, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Landi, M.; Brunetti, C.; Giordano, C.; Remorini, D.; Gould, K.S.; Guidi, L. Epidermal coumaroyl anthocyanins protect sweet basil against excess light stress: Multiple consequences of light attenuation. Physiol. Plant. 2014, 152, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Win, P.; Zhang, H.; Li, C.; Shen, Y.; He, F.; Luo, K. PtrARF2.1 Is Involved in Regulation of Leaf Development and Lignin Biosynthesis in Poplar Trees. Int. J. Mol. Sci. 2019, 20, 4141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, N.; Vanhaeren, H.; Inzé, D. Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Sun, N.; Zhang, F.; Yu, R.; Chen, H.; Deng, X.; Wei, N. SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis. Plant Cell 2020, 32, 3792–3811. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Xu, Z.H.; Xue, H.W. Arabidopsis membrane steroid binding protein 1 is involved in inhibition of cell elongation. Plant Cell 2005, 17, 116–131. [Google Scholar] [CrossRef] [Green Version]
- Bishop, G.J.; Koncz, C. Brassinosteroids and plant steroid hormone signaling. Plant Cell 2002, 14, S97–S110. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ori, N.; Cohen, A.R.; Etzioni, A.; Brand, A.; Yanai, O.; Shleizer, S.; Menda, N.; Amsellem, Z.; Efroni, I.; Pekker, I.; et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 2007, 39, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.P.; Furumizu, C.; Efroni, I.; Eshed, Y.; Bowman, J.L. Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 2016, 5, e15023. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Guan, C.; Jiao, Y. Molecular Mechanisms of Leaf Morphogenesis. Mol. Plant 2018, 11, 1117–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ercoli, M.F.; Rojas, A.M.; Debernardi, J.M.; Palatnik, J.F.; Rodriguez, R.E. Control of cell proliferation and elongation by miR396. Plant Signal. Behav. 2016, 11, e1184809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, S.; Ohashi, Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 2002, 30, 337–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2009, 15, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffrey, C.; Stebbins, G.L. Flowering Plants. Evolution above the Species Level. Kew Bull. 1977, 32, 273. [Google Scholar] [CrossRef]
- Juntheikki-Palovaara, I.; Tähtiharju, S.; Lan, T.; Broholm, S.K.; Rijpkema, A.S.; Ruonala, R.; Kale, L.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. 2014, 79, 783–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, R.; Peng, Y.; Zhao, Y.; Wang, X. Genome-Wide Identification and Expression Analysis of the TCP Genes in Jatropha curcas L. Reveals Its Roles in Involvement of Leaf Shape. Forests 2023, 14, 780. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, L.; Yung, W.S.; Su, W.; Huang, M. Ectopic expression of Torenia fournieri TCP8 and TCP13 alters the leaf and petal phenotypes in Arabidopsis thaliana. Physiol. Plant. 2021, 173, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, T.; Horiguchi, G.; Kim, G.T.; Ohgishi, M.; Sakai, T.; Tsukaya, H. The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol. 2005, 46, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.H.; Ko, J.H.; Lee, S.; Lee, Y.; Pak, J.H.; Kim, J.H. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009, 151, 655–668. [Google Scholar] [CrossRef] [Green Version]
- Mecchia, M.A.; Debernardi, J.M.; Rodriguez, R.E.; Schommer, C.; Palatnik, J.F. MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech. Dev. 2013, 130, 2–13. [Google Scholar] [CrossRef]
- Gonzalez, N.; De Bodt, S.; Sulpice, R.; Jikumaru, Y.; Chae, E.; Dhondt, S.; Van Daele, T.; De Milde, L.; Weigel, D.; Kamiya, Y.; et al. Increased leaf size: Different means to an end. Plant Physiol. 2010, 153, 1261–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A Rare Allele of GS2 Enhances Grain Size and Grain Yield in Rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casadevall, R.; Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F.; Casati, P. Repression of growth regulating factors by the microRNA396 inhibits cell proliferation by UV-B radiation in Arabidopsis leaves. Plant Cell 2013, 25, 3570–3583. [Google Scholar] [CrossRef] [Green Version]
- Fina, J.; Casadevall, R.; AbdElgawad, H.; Prinsen, E.; Markakis, M.N.; Beemster, G.T.S.; Casati, P. UV-B Inhibits Leaf Growth through Changes in Growth Regulating Factors and Gibberellin Levels. Plant Physiol. 2017, 174, 1110–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltramino, M.; Ercoli, M.F.; Debernardi, J.M.; Goldy, C.; Rojas, A.M.L.; Nota, F.; Alvarez, M.E.; Vercruyssen, L.; Inzé, D.; Palatnik, J.F.; et al. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 2018, 8, 13447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Zhang, D.; Xue, M.; Qian, J.; He, Y.; Wang, S. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 2014, 56, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zeng, J.; Liu, Q. The Rice miR396-GRF-GIF-SWI/SNF Module: A Player in GA Signaling. Front. Plant Sci. 2022, 12, 786641. [Google Scholar] [CrossRef] [PubMed]




| Gene | Amplified Fragment Length (bp) | Forward Primer (5′–3′) (Tm) | Reverse Primer (5′–3′) (Tm) |
|---|---|---|---|
| Fm5MAT | 205 | ACTTCCATCCAATTC AGCGTCT (53) | GTTAGAGAAACGGAG TCCCCAG (57) |
| Fm3GGT | 241 | GGCAACAACAACTTG TACCCAG (55) | ACCCAATCTTGGTTGC CAATTT (51) |
| FmGRF4 | 159 | AGGAACTGGAACAT CAAGCCAT (53) | ATTTCTTCCCATCTGTC CTCCG (55) |
| FmGRF7 | 121 | TAAATTGGCTGCAAA CATGGGG (53) | AAAGAGAAGATCAG CAGGCACA (53) |
| FmCYP75B1 | 225 | CGGAGCAGTTTCTTA AGGTCCA (55) | CTAAGGCGTGTGCAA GAATAGC (55) |
| FmC12RT1 | 231 | GCACATGGACATGTT TTTCCCT (53) | TGGCATAAGATTTGGT GGGACA (53) |
| FmLHCB6-1 | 230 | TGCATTCTTGAATGG TGGCAAG (53) | ATGCTGGATCCTTTCC AAGACC (55) |
| FmLHCA2 | 241 | TGATCCTGACAGACC ATTGTGG (55) | AGCAGTGTACCATGA AGGAGTG (55) |
| FmLHCB1 | 220 | AGATCACAGGCAAC GGAAGAAT (53) | ACCTCAAGCTCACGGT TCTTAG (55) |
| FmLHCB4 | 228 | ATTCGGAACCAAGA AAGCTCCA (53) | GTCCCAATAATATCAC CCCCA (52) |
| FmUGT79B1-1 | 178 | TGTTCAAAGCCAGAT TTCGTCG (53) | TATCGCACTGCGCAAT TGTTAG (53) |
| FmUGT79B1-2 | 241 | GGCAACAACAACTT GTACCCAG (55) | ACCCAATCTTGGTTGC CAATTT (51) |
| FmLHCB6-2 | 223 | AGCCAGATTTTGTTGT CTGCAC (53) | AGAATGCTGGATCCTT TCCGAG (55) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Yan, J.; Lu, H.; Su, W.; Gao, S.; Wang, Y.; Zhan, Y.; Zeng, F. Phenotypic Differences of Leaves and Transcriptome Analysis of Fraxinus mandshurica × Fraxinus sogdiana F1 Variety. Forests 2023, 14, 1554. https://doi.org/10.3390/f14081554
He L, Yan J, Lu H, Su W, Gao S, Wang Y, Zhan Y, Zeng F. Phenotypic Differences of Leaves and Transcriptome Analysis of Fraxinus mandshurica × Fraxinus sogdiana F1 Variety. Forests. 2023; 14(8):1554. https://doi.org/10.3390/f14081554
Chicago/Turabian StyleHe, Liming, Jialin Yan, Han Lu, Wenlong Su, Shangzhu Gao, Yubin Wang, Yaguang Zhan, and Fansuo Zeng. 2023. "Phenotypic Differences of Leaves and Transcriptome Analysis of Fraxinus mandshurica × Fraxinus sogdiana F1 Variety" Forests 14, no. 8: 1554. https://doi.org/10.3390/f14081554
APA StyleHe, L., Yan, J., Lu, H., Su, W., Gao, S., Wang, Y., Zhan, Y., & Zeng, F. (2023). Phenotypic Differences of Leaves and Transcriptome Analysis of Fraxinus mandshurica × Fraxinus sogdiana F1 Variety. Forests, 14(8), 1554. https://doi.org/10.3390/f14081554






