Excluding Roots or Mycorrhizal Hyphae Alters the Microbial Community and Function by Decreasing Available C and N in a Subtropical Chinese Fir Forest
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Experimental Design
2.3. Soil Sampling
2.4. Soil Analyses
2.5. Soil DNA Extraction and PCR Amplification
2.6. Bioinformatic Analysis
2.7. Statistical Analysis
3. Results
3.1. Soil Properties
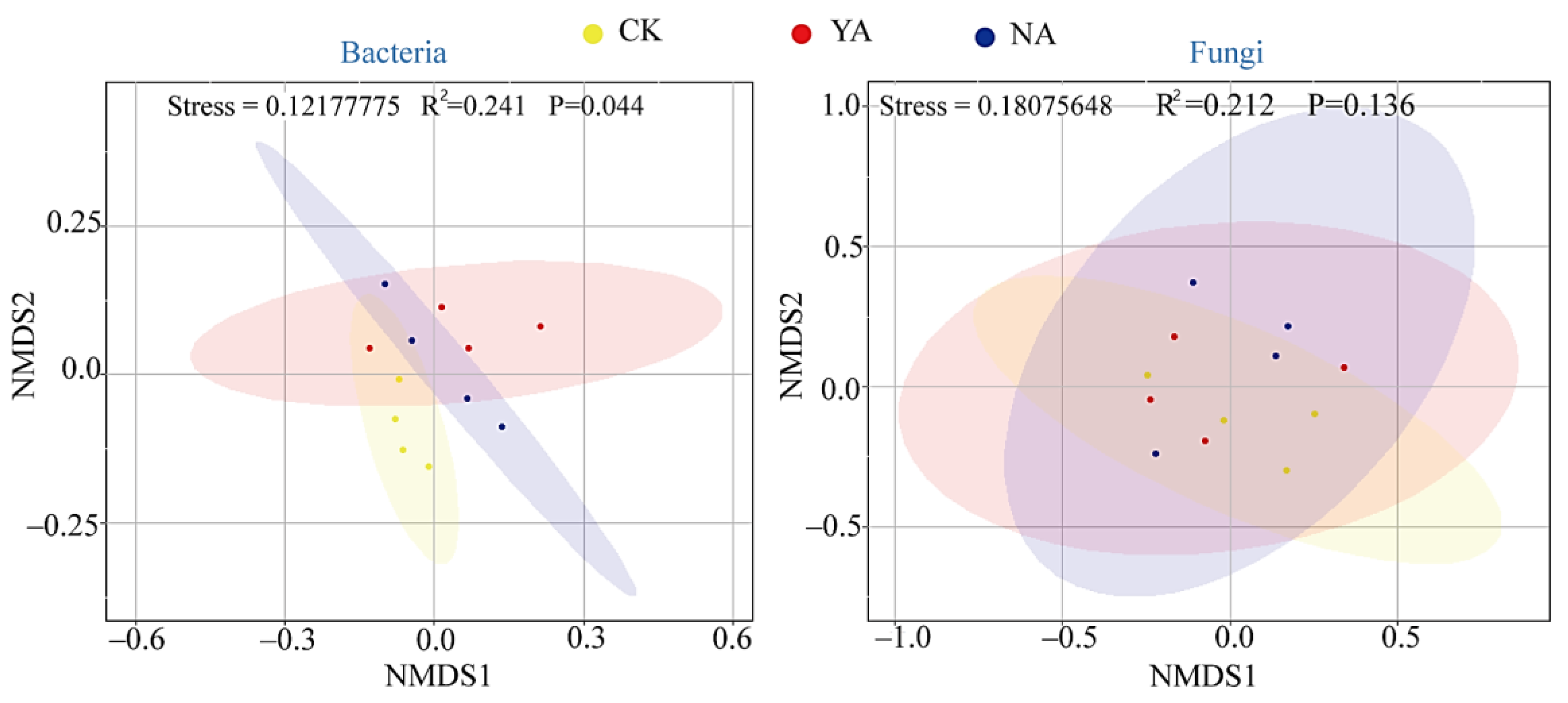
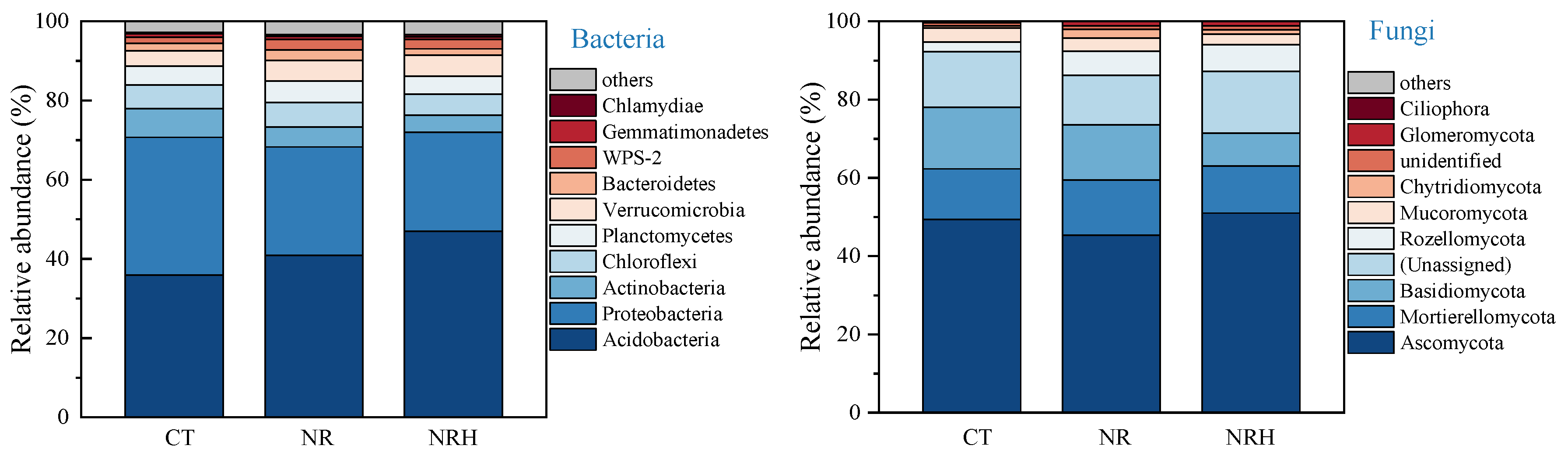
3.2. Microbial Diversity and Community Composition
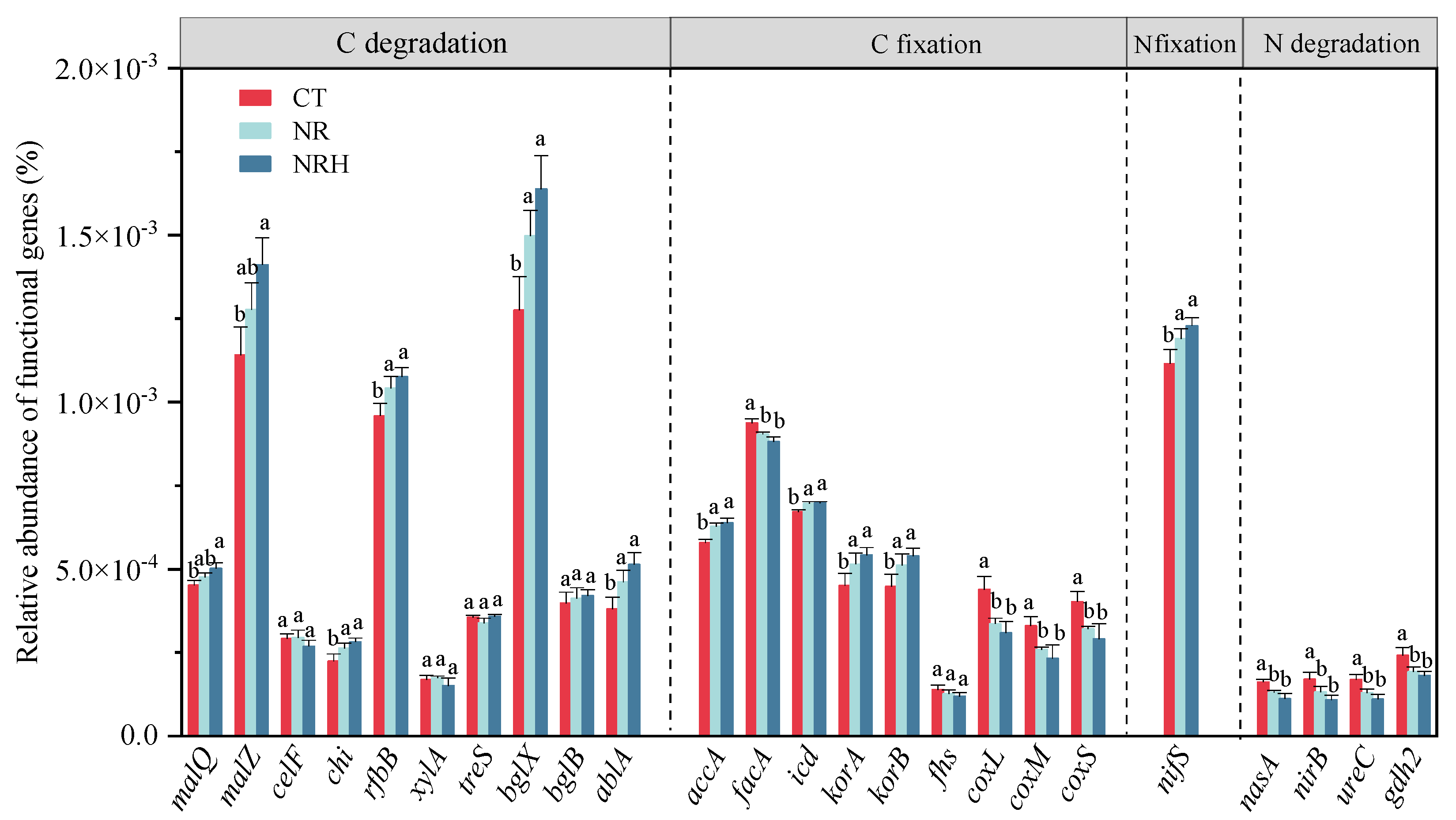
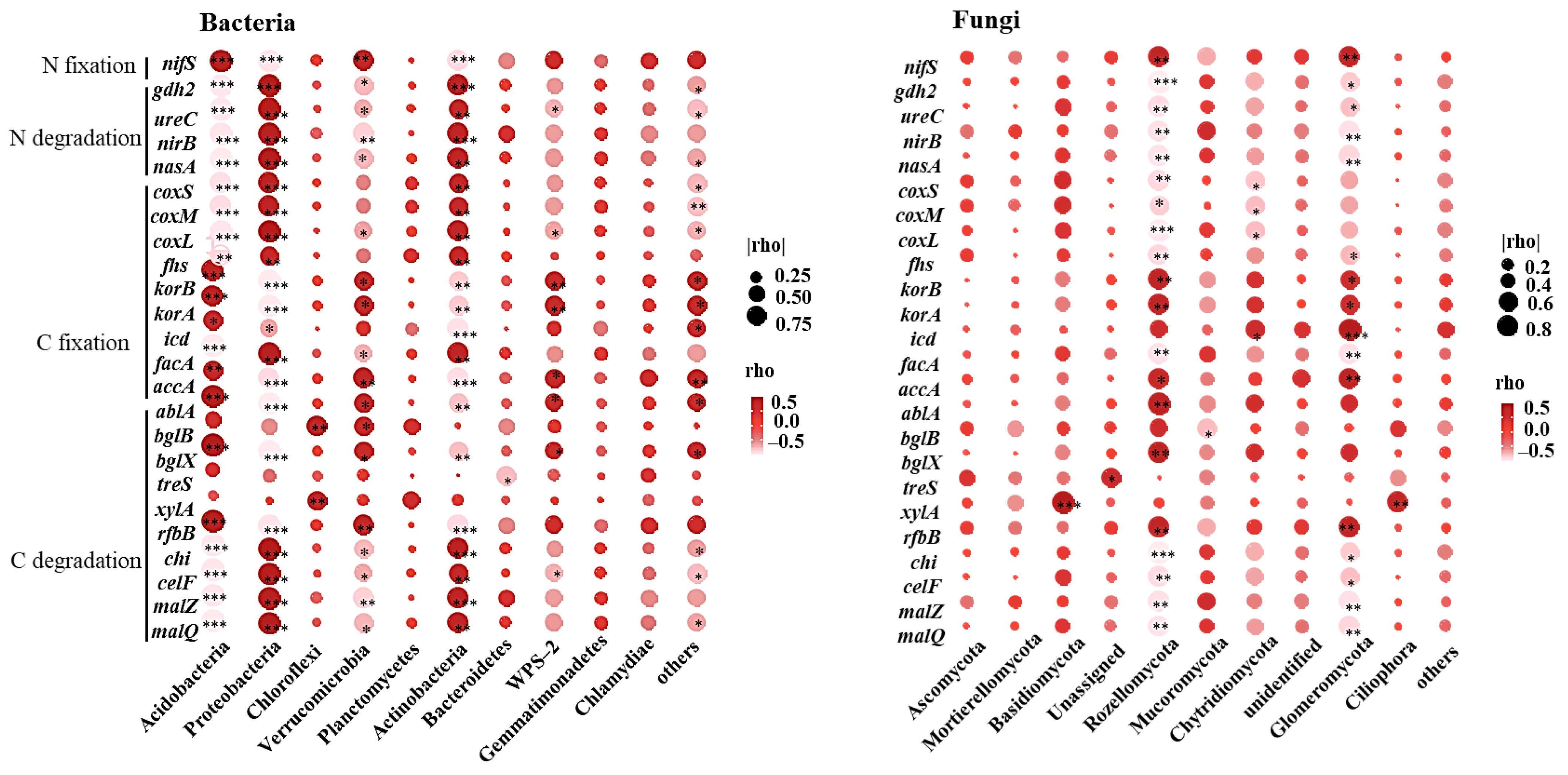
3.3. Microbial Functional Prediction
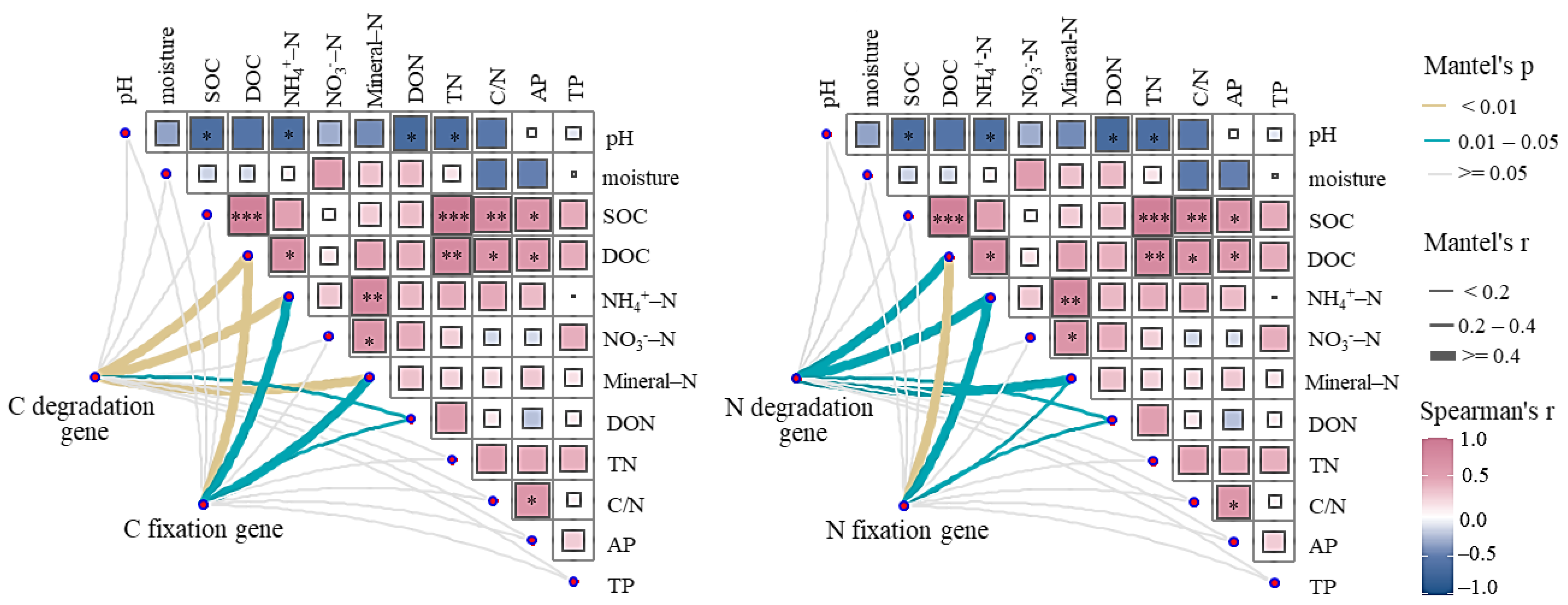
3.4. Influencing Factors of Microbial Community and Function
3.5. Relationships between Microbial Community Composition and Function
4. Discussions
4.1. Influence of Excluding Roots and Mycorrhizal Hyphae on Soil Properties
4.2. Influence of Excluding Roots and Mycorrhizal Hyphae on Microbial Diversity and Community Composition
4.3. Influence of Excluding Roots and Mycorrhizal Hyphae on Microbial Function in Meditating Soil C and N Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sokol, N.W.; Bradford, M.A. Microbial formation of stable soil carbon is more efficient from belowground than aboveground input. Nat. Geosci. 2019, 12, 46–53. [Google Scholar] [CrossRef]
- Crowther, T.W.; van den Hoogen, J.; Wan, J.; Mayes, M.A.; Keiser, A.D.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef]
- Mukhtar, H.; Wunderlich, R.F.; Muzaffar, A.; Ansari, A.; Shipin, O.V.; Cao, T.N.; Lin, Y.P. Soil microbiome feedback to climate change and options for mitigation. Sci. Total Environ. 2023, 882, 163412. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Moorhead, D.L.; Wang, X.; Xu, M.; Wang, X.; Wei, X.; Zhu, Z.; Ge, T.; Peng, S.; Zhu, B.; et al. Decreasing microbial phosphorus limitation increases soil carbon release. Geoderma 2022, 419, 115868. [Google Scholar] [CrossRef]
- Soong, J.L.; Fuchslueger, L.; Maranon-Jimenez, S.; Torn, M.S.; Janssens, I.A.; Penuelas, J.; Richter, A. Microbial carbon limitation: The need for integrating microorganisms into our understanding of ecosystem carbon cycling. Glob. Chang. Biol. 2020, 26, 1953–1961. [Google Scholar] [CrossRef] [PubMed]
- Panchal, P.; Preece, C.; Penuelas, J.; Giri, J. Soil carbon sequestration by root exudates. Trends Plant Sci. 2022, 27, 749–757. [Google Scholar] [CrossRef]
- Huang, J.; Liu, W.; Yang, S.; Yang, L.; Peng, Z.; Deng, M.; Xu, S.; Zhang, B.; Ahirwal, J.; Liu, L. Plant carbon inputs through shoot, root, and mycorrhizal pathways affect soil organic carbon turnover differently. Soil Biol. Biochem. 2021, 160, 108332. [Google Scholar] [CrossRef]
- Prescott, C.E.; Grayston, S.J.; Helmisaari, H.S.; Kastovska, E.; Korner, C.; Lambers, H.; Meier, I.C.; Millard, P.; Ostonen, I. Surplus carbon drives allocation and plant-soil interactions. Trends Ecol. Evol. 2020, 35, 1110–1118. [Google Scholar] [CrossRef]
- Williams, A.; de Vries, F.T. Plant root exudation under drought: Implications for ecosystem functioning. New Phytol. 2020, 225, 1899–1905. [Google Scholar] [CrossRef]
- Freeman, C.; Fenner, N.; Ostle, N.; Kang, H.; Dowrick, D.; Reynolds, B.; Lock, M.; Sleep, D.; Hughes, S.; Hudson, J. Export of dissolved organic carbon from peatlands under elevated carbon dioxide levels. Nature 2004, 430, 195–198. [Google Scholar] [CrossRef]
- Zhao, C.; He, X.; Dan, X.; He, M.; Zhao, J.; Meng, H.; Cai, Z.; Zhang, J. Soil dissolved organic matters mediate bacterial taxa to enhance nitrification rates under wheat cultivation. Sci. Total Environ. 2022, 828, 154418. [Google Scholar] [CrossRef]
- Ma, S.; Zhu, W.; Wang, W.; Li, X.; Sheng, Z. Microbial assemblies with distinct trophic strategies drive changes in soil microbial carbon use efficiency along vegetation primary succession in a glacier retreat area of the southeastern Tibetan Plateau. Sci. Total Environ. 2023, 867, 161587. [Google Scholar] [CrossRef]
- Bastida, F.; Eldridge, D.J.; Garcia, C.; Kenny Png, G.; Bardgett, R.D.; Delgado-Baquerizo, M. Soil microbial diversity-biomass relationships are driven by soil carbon content across global biomes. ISME 2021, 15, 2081–2091. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhong, X.; Xia, Y.; Xu, L.; Zeng, Q.; Yang, L.; Fan, Y. Long-term nitrogen addition exerts minor effects on microbial community but alters sensitive microbial species in a subtropical natural forest. Forests 2023, 14, 928. [Google Scholar] [CrossRef]
- Hammarlund, S.P.; Harcombe, W.R. Refining the stress gradient hypothesis in a microbial community. Proc. Natl. Acad. Sci. USA 2019, 116, 15760–15762. [Google Scholar] [CrossRef]
- Wang, C.; Liu, D.; Bai, E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Pausch, J.; Kuzyakov, Y. Carbon input by roots into the soil: Quantification of rhizodeposition from root to ecosystem scale. Glob. Chang. Biol. 2018, 24, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-Gonzalez, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Wang, J.; Shi, X.; Zheng, C.; Suter, H.; Huang, Z. Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci. Total Environ. 2021, 755, 142449. [Google Scholar] [CrossRef]
- Roller, B.R.; Schmidt, T.M. The physiology and ecological implications of efficient growth. ISME 2015, 9, 1481–1487. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Isobe, K.; Ise, Y.; Kato, H.; Oda, T.; Vincenot, C.E.; Koba, K.; Tateno, R.; Senoo, K.; Ohte, N. Consequences of microbial diversity in forest nitrogen cycling: Diverse ammonifiers and specialized ammonia oxidizers. ISME 2020, 14, 12–25. [Google Scholar] [CrossRef]
- Bauer, S.; Vasu, P.; Persson, S.; Mort, A.J.; Somerville, C.R. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. USA 2006, 103, 11417–11422. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Li, J.; Yan, B.; Wei, F.; Wang, G.; Liu, G. The effects of nitrogen addition on soil organic carbon decomposition and microbial C-degradation functional genes abundance in a Pinus tabulaeformis forest. Forest Ecol. Manag. 2021, 489, 119098. [Google Scholar] [CrossRef]
- Kelly, C.N.; Schwaner, G.W.; Cumming, J.R.; Driscoll, T.P. Metagenomic reconstruction of nitrogen and carbon cycling pathways in forest soil: Influence of different hardwood tree species. Soil Biol. Biochem. 2021, 156, 108226. [Google Scholar] [CrossRef]
- Liao, H.; Hao, X.; Qin, F.; Delgado-Baquerizo, M.; Liu, Y.; Zhou, J.; Cai, P.; Chen, W.; Huang, Q. Microbial autotrophy explains large-scale soil CO2 fixation. Glob. Chang. Biol. 2023, 29, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Piao, S.; Fang, J.; Ciais, P.; Peylin, P.; Huang, Y.; Sitch, S.; Wang, T. The carbon balance of terrestrial ecosystems in China. Nature 2009, 458, 1009–1013. [Google Scholar] [CrossRef]
- Jia, S.; Liu, X.; Lin, W.; Li, X.; Yang, L.; Sun, S.; Hui, D.; Guo, J.; Zou, X.; Yang, Y. Tree roots exert greater influence on soil microbial necromass carbon than above-ground litter in subtropical natural and plantation forests. Soil Biol. Biochem. 2022, 173, 108811. [Google Scholar] [CrossRef]
- Feng, J.; He, K.; Zhang, Q.; Han, M.; Zhu, B. Changes in plant inputs alter soil carbon and microbial communities in forest ecosystems. Glob. Chang. Biol. 2022, 28, 3426–3440. [Google Scholar] [CrossRef]
- Sun, L.; Ataka, M.; Kominami, Y.; Yoshimura, K. Relationship between fine-root exudation and respiration of two Quercus species in a Japanese temperate forest. Tree Physiol. 2017, 37, 1011–1020. [Google Scholar] [CrossRef]
- Shahzad, T.; Chenu, C.; Genet, P.; Barot, S.; Perveen, N.; Mougin, C.; Fontaine, S. Contribution of exudates, arbuscular mycorrhizal fungi and litter depositions to the rhizosphere priming effect induced by grassland species. Soil Biol. Biochem. 2015, 80, 146–155. [Google Scholar] [CrossRef]
- De Graaff, M.A.; Classen, A.T.; Castro, H.F.; Schadt, C.W. Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol. 2010, 188, 1055–1064. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Neilson, J.W.; Kushwaha, P.; Maier, R.M.; Barberan, A. Life-history strategies of soil microbial communities in an arid ecosystem. ISME 2021, 15, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Coskun, D.; Britto, D.T.; Shi, W.; Kronzucker, H.J. How plant root exudates shape the nitrogen cycle. Trends Plant Sci. 2017, 22, 661–673. [Google Scholar] [CrossRef]
- Liu, Y.; Evans, S.E.; Friesen, M.L.; Tiemann, L.K. Root exudates shift how N mineralization and N fixation contribute to the plant-available N supply in low fertility soils. Soil Biol. Biochem. 2022, 165, 108541. [Google Scholar] [CrossRef]
- Lloyd, J.R.; Blennow, A.; Burhenne, K.; Kossmann, J. Repression of a novel isoform of disproportionating enzyme (stDPE2) in potato leads to inhibition of starch degradation in leaves but not tubers stored at low temperature. Plant Physiol. 2004, 134, 1347–1354. [Google Scholar] [CrossRef][Green Version]
- Radakovits, R.; Jinkerson, R.E.; Fuerstenberg, S.I.; Tae, H.; Settlage, R.E.; Boore, J.L.; Posewitz, M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 2012, 3, 686. [Google Scholar] [CrossRef] [PubMed]
- Boles, E.; Lehnert, W.; Zimmermann, F.K. The role of the NAD-dependent glutamate dehydrogenase in restoring growth on glucose of a Saccharomyces cerevisiae phosphoglucose isomerase mutant. Eur. J. Biochem. 1993, 217, 469–477. [Google Scholar] [CrossRef]
- Harborne, N.R.; Griffiths, L.; Busby, S.J.; Cole, J.A. Transcriptional control, translation and function of the products of the five open reading frames of the Escherichia coli nir operon. Mol. Microbiol. 1992, 6, 2805–2813. [Google Scholar] [CrossRef]
- Curatti, L.; Rubio, L.M. Challenges to develop nitrogen-fixing cereals by direct nif-gene transfer. Plant Sci. 2014, 225, 130–137. [Google Scholar] [CrossRef] [PubMed]

| Soil Properties | Treatment | ||
|---|---|---|---|
| CT | NR | NRH | |
| pH | 4.34 ± 0.05 a | 4.41 ± 0.05 a | 4.4 ± 0.04 ba |
| Moisture | 0.25 ± 0.02 a | 0.23 ± 0.03 a | 0.24 ± 0.02 a |
| SOC (g·kg−1) | 21.34 ± 2.06 a | 17.89 ± 2.62 a | 17.05 ± 4.83 a |
| DOC (mg·kg−1) | 39.43 ± 10.71 a | 29.03 ± 7.78 ab | 25.53 ± 4.26 b |
| NH4+-N (mg·kg−1) | 12.68 ± 4.51 a | 6.27 ± 1.93 b | 5.1 ± 0.88 b |
| NO3−-N (mg·kg−1) | 5.32 ± 0.92 a | 4.45 ± 0.8 ab | 3.37 ± 0.63 b |
| Mineral N (mg·kg−1) | 17.99 ± 4.57 a | 11.3 ± 0.87 b | 9.2 ± 1.9 b |
| DON (mg·kg−1) | 0.92 ± 0.37 a | 0.7 ± 0.36 ab | 0.37 ± 0.08 b |
| TN (g·kg−1) | 1.41 ± 0.13 a | 1.24 ± 0.14 a | 1.2 ± 0.19 a |
| C/N | 15.17 ± 0.6 a | 14.41 ± 0.99 a | 13.98 ± 2.09 a |
| AP (mg·kg−1) | 2.48 ± 0.54 a | 2 ± 0.86 a | 2.04 ± 0.78 a |
| TP (g·kg−1) | 0.15 ± 0.01 a | 0.15 ± 0.03 a | 0.14 ± 0.01 a |
| Microbial Diversity | Treatment | |||
|---|---|---|---|---|
| CT | NR | NRH | ||
| Bacteria | richness | 3607 ± 127 a | 3298 ± 87 b | 3158 ± 71 b |
| chao1 | 3609 ± 127 a | 3299 ± 87 b | 3159 ± 71 b | |
| simpson | 0.0056 ± 0.0005 b | 0.0057 ± 0.0006 ab | 0.0068 ± 0.0007 a | |
| shannon | 3 ± 0.02 a | 3 ± 0.03 a | 3 ± 0.03 a | |
| Fungi | richness | 1380 ± 104 a | 1147 ± 27 b | 1059 ± 79 b |
| chao1 | 1381 ± 104 a | 1148 ± 27 b | 1060 ± 79 b | |
| simpson | 0.04 ± 0.032 a | 0.027 ± 0.003 a | 0.021 ± 0.014 a | |
| shannon | 2 ± 0.15 a | 2 ± 0.05 a | 2 ± 0.11 a | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, P.; Xu, L.; Yue, K.; Yang, L. Excluding Roots or Mycorrhizal Hyphae Alters the Microbial Community and Function by Decreasing Available C and N in a Subtropical Chinese Fir Forest. Forests 2023, 14, 1847. https://doi.org/10.3390/f14091847
Lian P, Xu L, Yue K, Yang L. Excluding Roots or Mycorrhizal Hyphae Alters the Microbial Community and Function by Decreasing Available C and N in a Subtropical Chinese Fir Forest. Forests. 2023; 14(9):1847. https://doi.org/10.3390/f14091847
Chicago/Turabian StyleLian, Pingping, Linglin Xu, Kai Yue, and Liuming Yang. 2023. "Excluding Roots or Mycorrhizal Hyphae Alters the Microbial Community and Function by Decreasing Available C and N in a Subtropical Chinese Fir Forest" Forests 14, no. 9: 1847. https://doi.org/10.3390/f14091847
APA StyleLian, P., Xu, L., Yue, K., & Yang, L. (2023). Excluding Roots or Mycorrhizal Hyphae Alters the Microbial Community and Function by Decreasing Available C and N in a Subtropical Chinese Fir Forest. Forests, 14(9), 1847. https://doi.org/10.3390/f14091847








