Individual Tree Species Identification for Complex Coniferous and Broad-Leaved Mixed Forests Based on Deep Learning Combined with UAV LiDAR Data and RGB Images
Abstract
:1. Introduction
2. Study Area and Data Acquisition
2.1. Study Area
2.2. Data Acquisition
2.2.1. UAV Data Acquisition
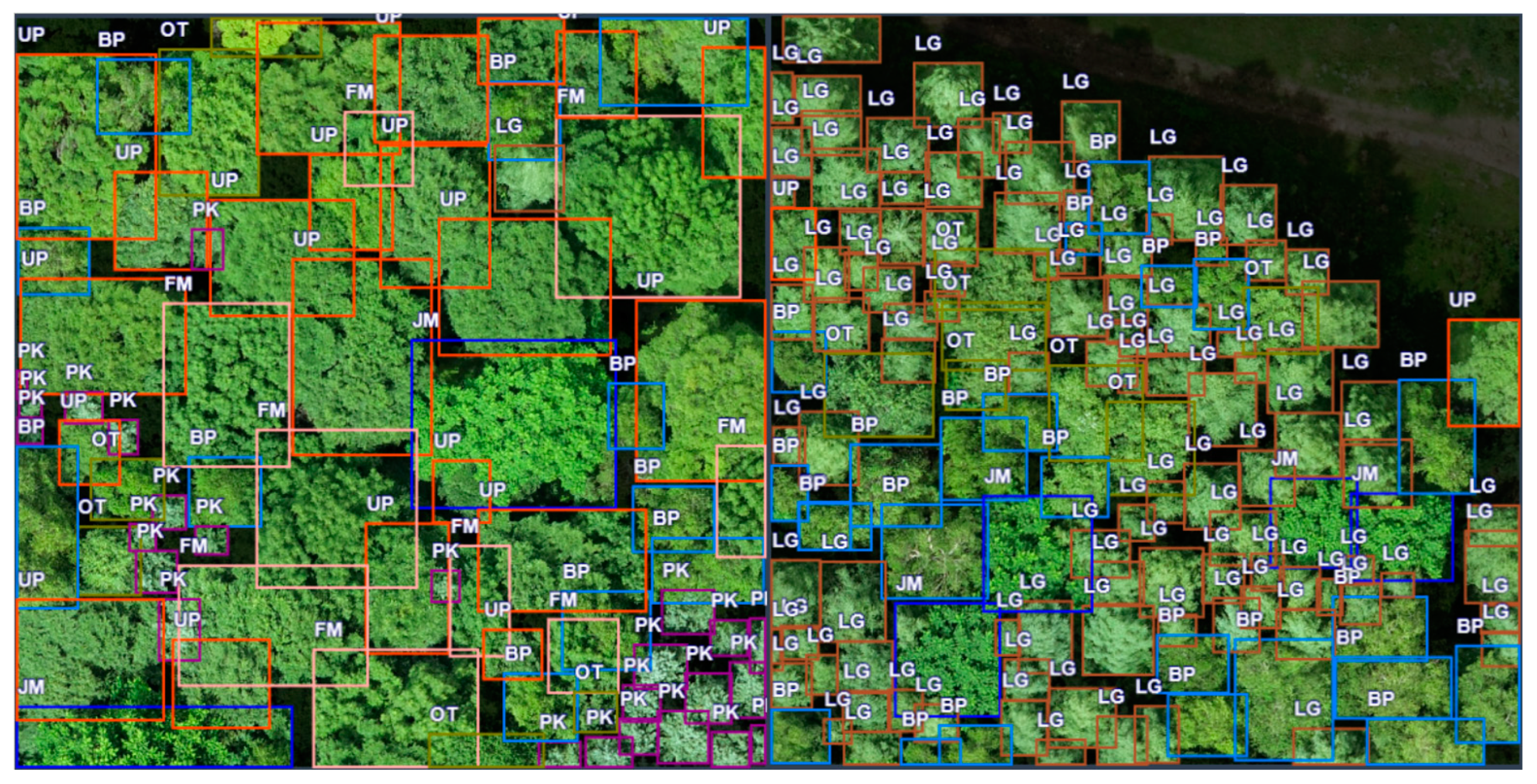
2.2.2. Plot Survey
3. Methods
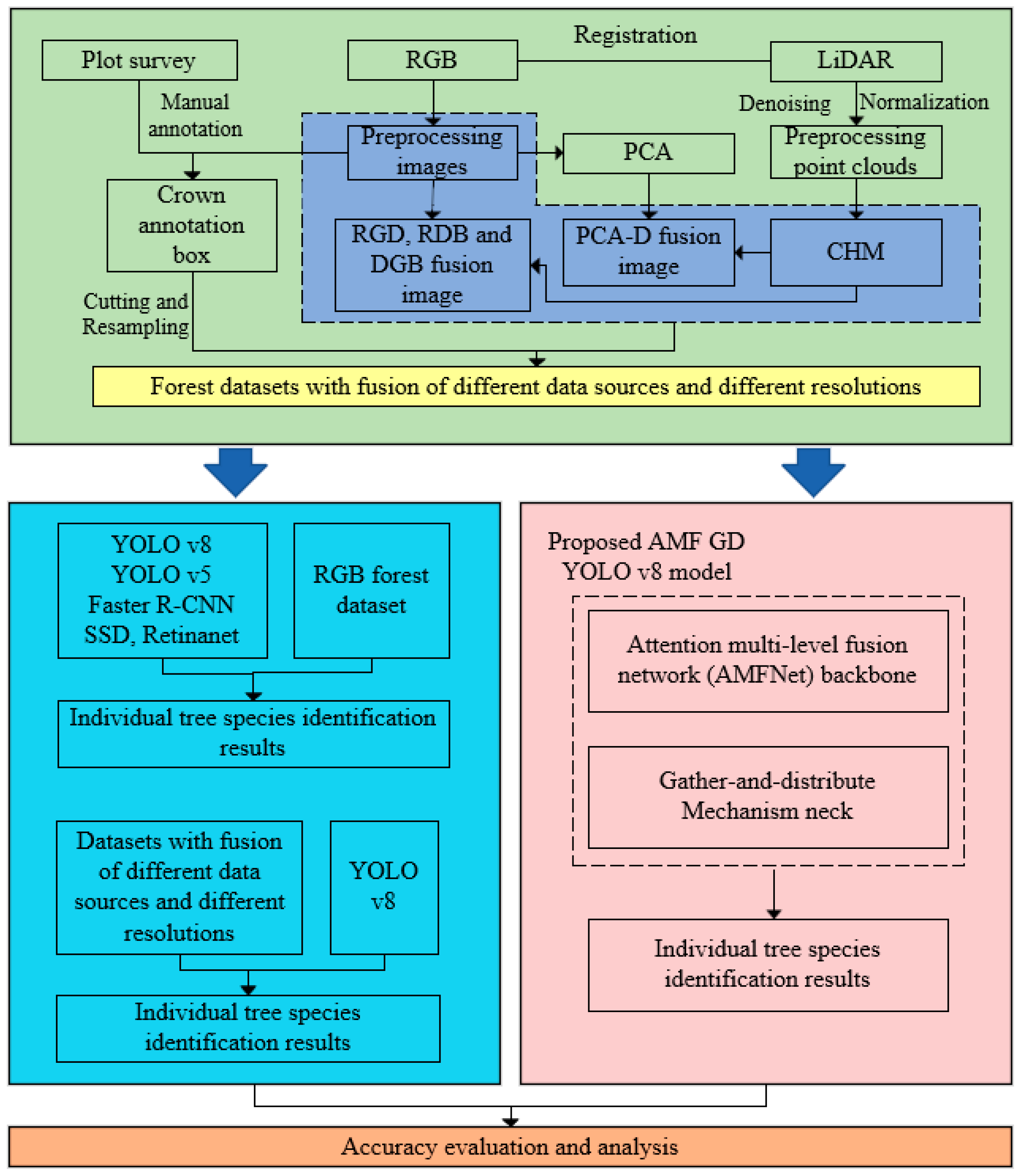
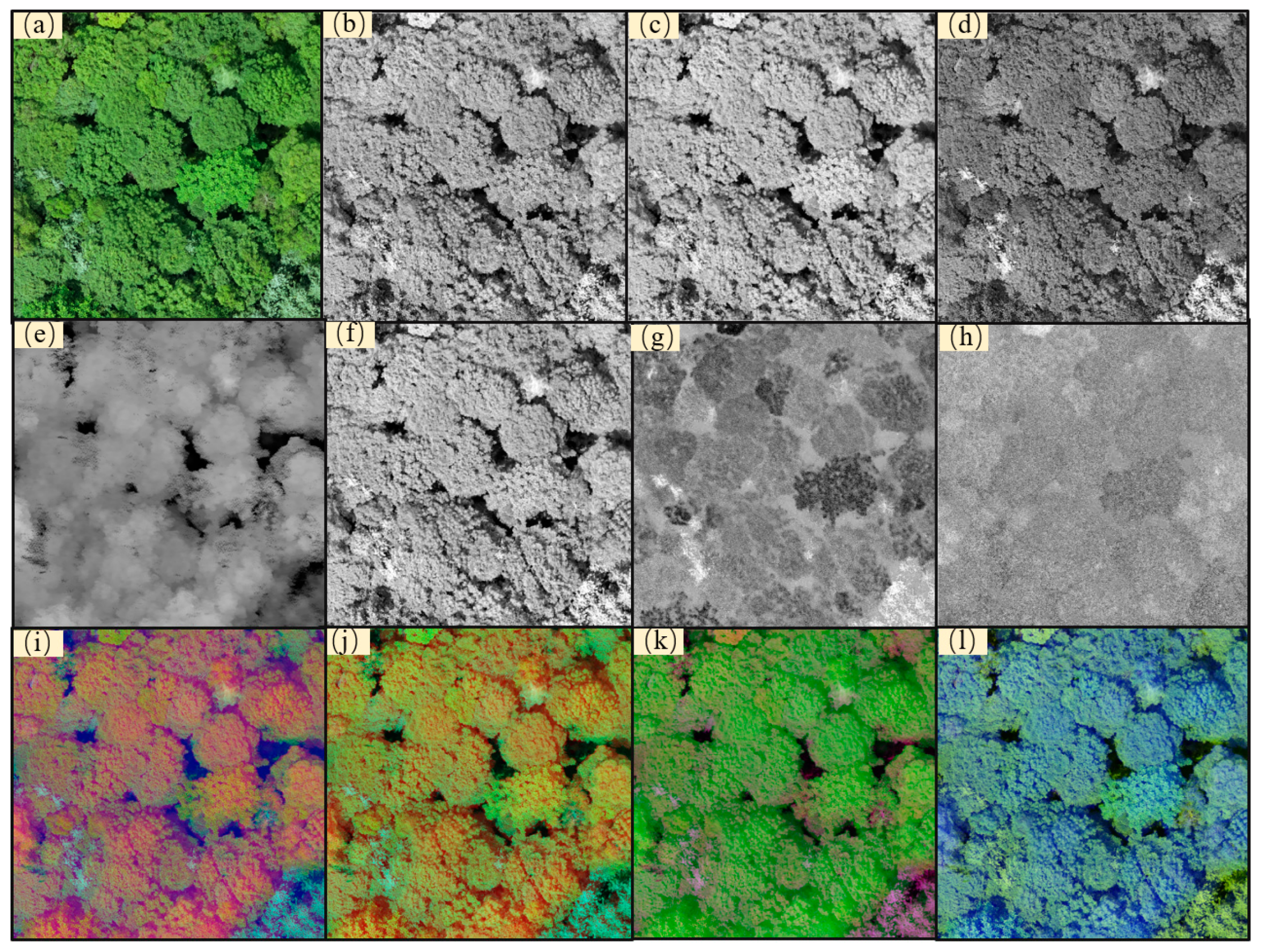
3.1. Data Preprocessing
3.2. Dataset Creation and Data Fusion
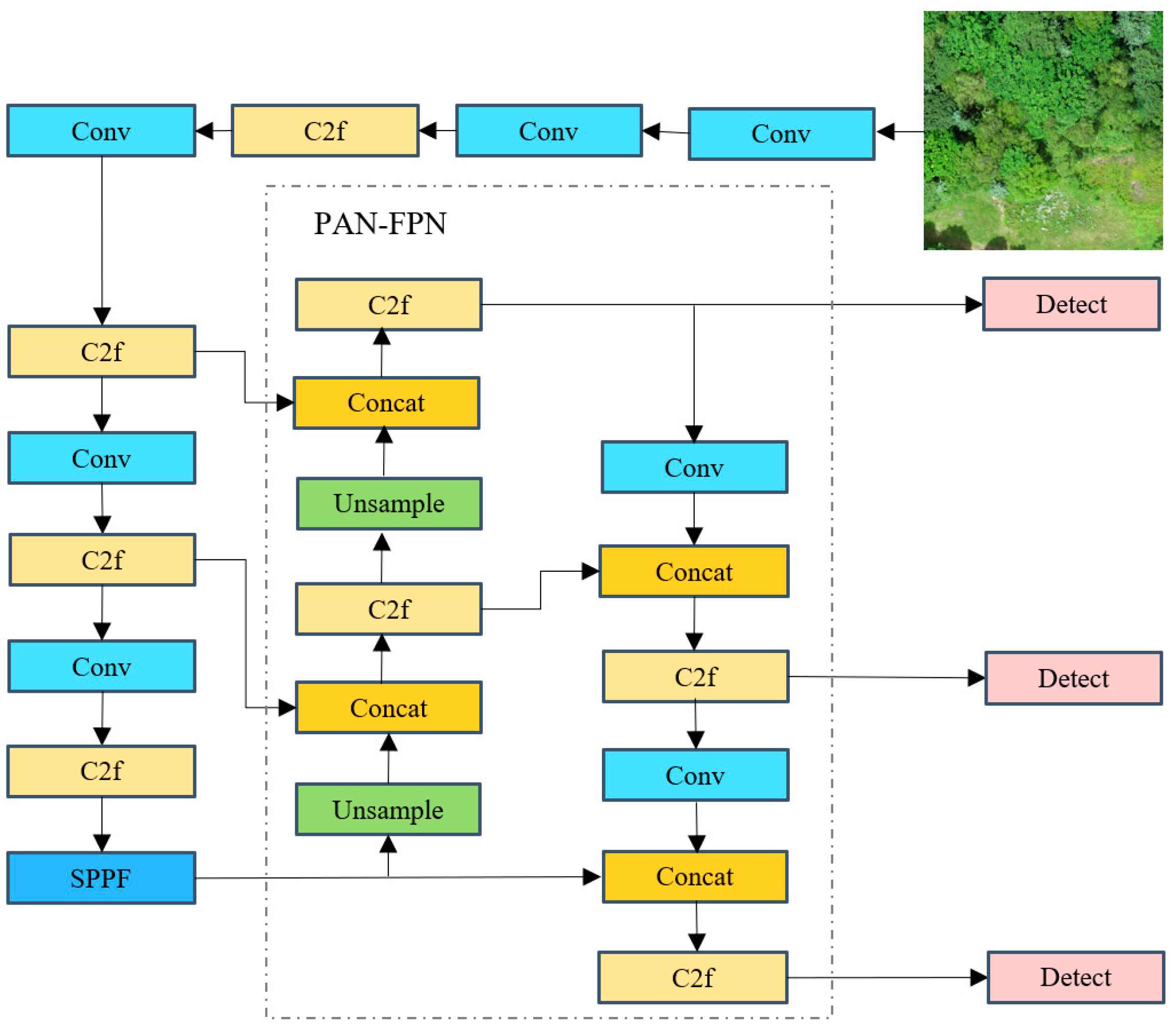
3.3. Performance Comparison of Different Object Detection Models
3.4. Tree Species Identification Effectiveness of Different Scales and Spatial Resolutions in YOLO v8
3.5. Tree Species Identification Performance of Different Data Fusion Methods
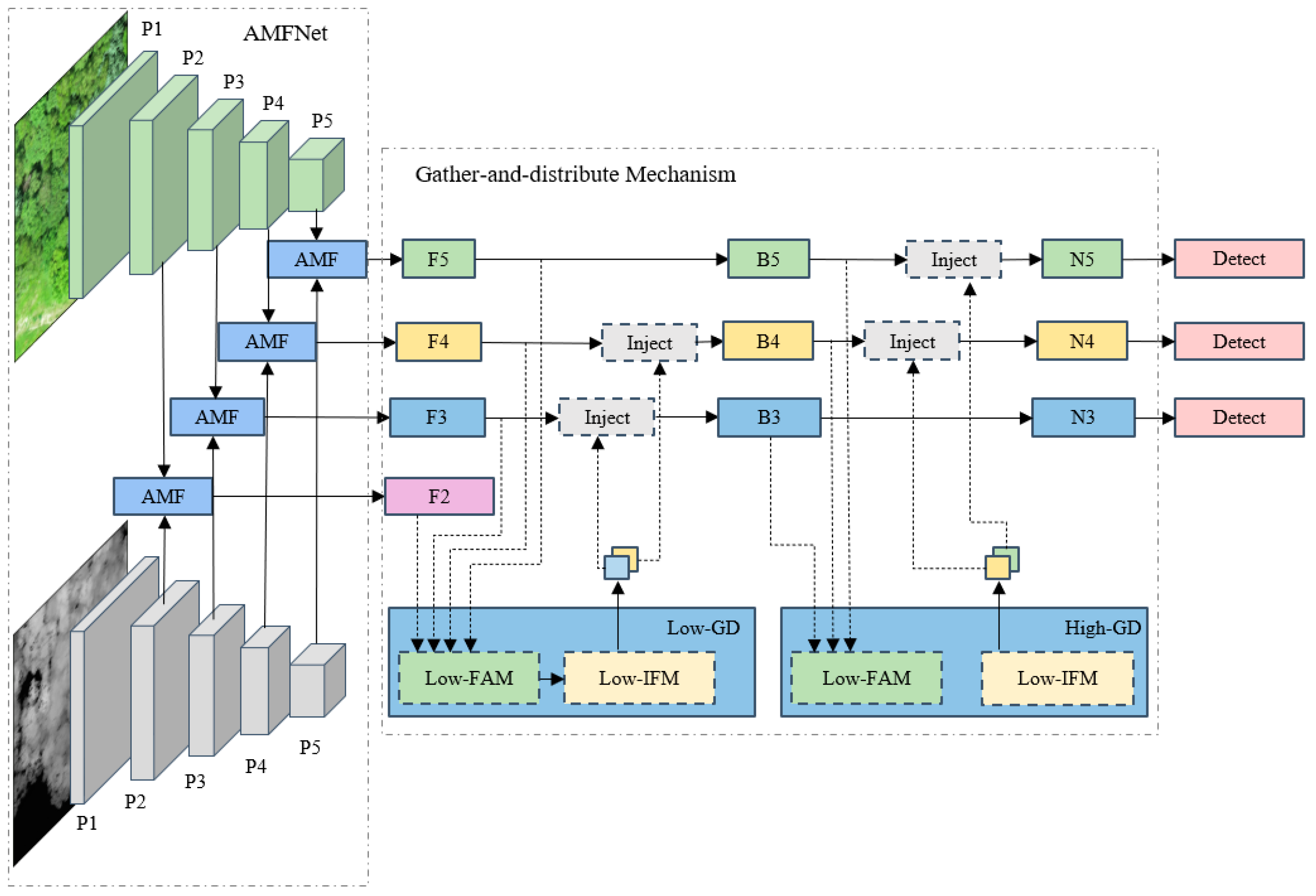
3.6. AMF GD YOLO v8 Model
3.6.1. AMFNet
3.6.2. Gather-and-Distribute Mechanism
3.7. Accuracy Evaluation and Experimental Environment
4. Results and Analysis
4.1. Individual Tree Species Identification Results of Different Models
4.2. Impact of Different Spatial Resolutions and YOLO v8 Scales on Individual Tree Species Identification
4.3. Tree Species Identification Results of Different Data Fusion Methods
4.4. AMF GD YOLO v8 Model Tree Species Identification Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.A. Identifying the genus or species of individual trees using a three-wavelength airborne lidar system. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Sterenczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Braga, J.R.G.; Peripato, V.; Dalagnol, R.; Ferreira, M.P.; Tarabalka, Y.; Aragao, L.; Velho, H.E.D.; Shiguemori, E.H.; Wagner, F.H. Tree crown delineation algorithm based on a convolutional neural network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef]
- de Almeida, D.R.A.; Broadbent, E.N.; Ferreira, M.P.; Meli, P.; Zambrano, A.M.A.; Gorgens, E.B.; Resende, A.F.; de Almeida, C.T.; do Amaral, C.H.; Corte, A.P.D.; et al. Monitoring restored tropical forest diversity and structure through UAV-borne hyperspectral and lidar fusion. Remote Sens. Environ. 2021, 264, 112582. [Google Scholar] [CrossRef]
- Terryn, L.; Calders, K.; Bartholomeus, H.; Bartolo, R.E.; Brede, B.; D’Hont, B.; Disney, M.; Herold, M.; Lau, A.; Shenkin, A.; et al. Quantifying tropical forest structure through terrestrial and UAV laser scanning fusion in Australian rainforests. Remote Sens. Environ. 2022, 271, 112912. [Google Scholar] [CrossRef]
- Li, Y.B.; Chai, G.Q.; Wang, Y.T.; Lei, L.T.; Zhang, X.L. ACE R-CNN: An attention complementary and edge detection-based instance segmentation algorithm for individual tree species identification using UAV RGB images and LiDAR data. Remote Sens. 2022, 14, 3035. [Google Scholar] [CrossRef]
- Shen, X.; Cao, L. Tree-species classification in subtropical forests using airborne hyperspectral and LiDAR data. Remote Sens. 2017, 9, 1180. [Google Scholar] [CrossRef]
- Zhao, D.; Pang, Y.; Liu, L.; Li, Z. Individual tree classification using airborne LiDAR and hyperspectral data in a natural mixed forest of Northeast China. Forests 2020, 11, 303. [Google Scholar] [CrossRef]
- Qin, H.M.; Zhou, W.Q.; Yao, Y.; Wang, W.M. Individual tree segmentation and tree species classification in subtropical broadleaf forests using UAV-based LiDAR, hyperspectral, and ultrahigh-resolution RGB data. Remote Sens. Environ. 2022, 280, 113143. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Evans, J.S.; Martinuzzi, S.; Gessler, P.E.; Hudak, A.T. Characterizing forest succession with lidar data: An evaluation for the Inland Northwest, USA. Remote Sens. Environ. 2009, 113, 946–956. [Google Scholar] [CrossRef]
- Lu, X.C.; Guo, Q.H.; Li, W.K.; Flanagan, J. A bottom-up approach to segment individual deciduous trees using leaf-off lidar point cloud data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 1–12. [Google Scholar] [CrossRef]
- Jaskierniak, D.; Lucieer, A.; Kuczera, G.; Turner, D.; Lane, P.N.J.; Benyon, R.G.; Haydon, S. Individual tree detection and crown delineation from Unmanned Aircraft System (UAS) LiDAR in structurally complex mixed species eucalypt forests. ISPRS J. Photogramm. Remote Sens. 2021, 171, 171–187. [Google Scholar] [CrossRef]
- Liu, B.J.; Hao, Y.S.; Huang, H.G.; Chen, S.X.; Li, Z.Y.; Chen, E.X.; Tian, X.; Ren, M. TSCMDL: Multimodal deep learning framework for classifying tree species using fusion of 2-D and 3-D features. IEEE Trans. Geosci. Remote Sens. 2023, 61, 4402711. [Google Scholar] [CrossRef]
- You, H.T.; Tang, X.; You, Q.X.; Liu, Y.; Chen, J.J.; Wang, F. Study on the differences between the extraction results of the structural parameters of individual trees for different tree species based on UAV LiDAR and high-resolution RGB images. Drones 2023, 7, 317. [Google Scholar] [CrossRef]
- Lombardi, E.; Rodríguez-Puerta, F.; Santini, F.; Chambel, M.R.; Climent, J.; de Dios, V.R.; Voltas, J. UAV-LiDAR and RGB imagery reveal large intraspecific variation in tree-level morphometric traits across different pine species evaluated in common gardens. Remote Sens. 2022, 14, 5904. [Google Scholar] [CrossRef]
- Deng, S.Q.; Katoh, M.; Yu, X.W.; Hyyppä, J.; Gao, T. Comparison of tree species classifications at the individual tree level by combining ALS data and RGB images using different algorithms. Remote Sens. 2016, 8, 1034. [Google Scholar] [CrossRef]
- Mayra, J.; Keski-Saari, S.; Kivinen, S.; Tanhuanpaa, T.; Hurskainen, P.; Kullberg, P.; Poikolainen, L.; Viinikka, A.; Tuominen, S.; Kumpula, T.; et al. Tree species classification from airborne hyperspectral and LiDAR data using 3D convolutional neural networks. Remote Sens. Environ. 2021, 256, 112322. [Google Scholar] [CrossRef]
- Hamraz, H.; Jacobs, N.B.; Contreras, M.A.; Clark, C.H. Deep learning for conifer/deciduous classification of airborne LiDAR 3D point clouds representing individual trees. ISPRS J. Photogramm. Remote Sens. 2019, 158, 219–230. [Google Scholar] [CrossRef]
- Liu, L.; Lim, S.; Shen, X.S.; Yebra, M. A hybrid method for segmenting individual trees from airborne lidar data. Comput. Electron. Agric. 2019, 163, 104871. [Google Scholar] [CrossRef]
- Roeder, M.; Latifi, H.; Hill, S.; Wild, J.; Svoboda, M.; Bruna, J.; Macek, M.; Novakova, M.H.; Guelch, E.; Heurich, M. Application of optical unmanned aerial vehicle-based imagery for the inventory of natural regeneration and standing deadwood in post-disturbed spruce forests. Int. J. Remote Sens. 2018, 39, 5288–5309. [Google Scholar] [CrossRef]
- Ferraz, A.; Bretar, F.; Jacquemoud, S.; Gonçalves, G.; Pereira, L.; Tomé, M.; Soares, P. 3-D mapping of a multi-layered Mediterranean forest using ALS data. Remote Sens. Environ. 2012, 121, 210–223. [Google Scholar] [CrossRef]
- Lee, H.; Slatton, K.C.; Roth, B.E.; Cropper, W.P. Adaptive clustering of airborne LiDAR data to segment individual tree crowns in managed pine forests. Int. J. Remote Sens. 2010, 31, 117–139. [Google Scholar] [CrossRef]
- Modzelewska, A.; Fassnacht, F.E.; Sterenczak, K. Tree species identification within an extensive forest area with diverse management regimes using airborne hyperspectral data. Int. J. Appl Earth Obs. Geoinf. 2020, 84, 101960. [Google Scholar] [CrossRef]
- Rana, P.; St-Onge, B.; Prieur, J.F.; Budei, B.C.; Tolvanen, A.; Tokola, T. Effect of feature standardization on reducing the requirements of field samples for individual tree species classification using ALS data. ISPRS J. Photogramm. Remote Sens. 2022, 184, 189–202. [Google Scholar] [CrossRef]
- Ke, Y.H.; Quackenbush, L.J. A review of methods for automatic individual tree-crown detection and delineation from passive remote sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Hoeser, T.; Kuenzer, C. Object detection and image segmentation with deep learning on earth observation data: A review-part I: Evolution and recent trends. Remote Sens. 2020, 12, 1667. [Google Scholar] [CrossRef]
- Hao, Z.B.; Lin, L.L.; Post, C.J.; Mikhailova, E.A.; Li, M.H.; Yu, K.Y.; Liu, J.; Chen, Y. Automated tree-crown and height detection in a young forest plantation using mask region-based convolutional neural network (Mask R-CNN). ISPRS J. Photogramm. Remote Sens. 2021, 178, 112–123. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.B.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 1137–1149. [Google Scholar] [CrossRef]
- Luo, M.; Tian, Y.A.; Zhang, S.W.; Huang, L.; Wang, H.Q.; Liu, Z.Q.; Yang, L. Individual tree detection in coal mine afforestation area based on improved Faster RCNN in UAV RGB images. Remote Sens. 2022, 14, 5545. [Google Scholar] [CrossRef]
- Xia, K.; Wang, H.; Yang, Y.H.; Du, X.C.; Feng, H.L. Automatic detection and parameter estimation of Ginkgo biloba in urban environment based on RGB Images. J. Sens. 2021, 2021, 6668934. [Google Scholar] [CrossRef]
- Beloiu, M.; Heinzmann, L.; Rehush, N.; Gessler, A.; Griess, V.C. Individual tree-crown detection and species identification in heterogeneous forests using aerial RGB imagery and deep learning. Remote Sens. 2023, 15, 1463. [Google Scholar] [CrossRef]
- Gan, Y.; Wang, Q.; Iio, A. Tree crown detection and delineation in a temperate deciduous forest from UAV RGB imagery using deep learning approaches: Effects of spatial resolution and species characteristics. Remote Sens. 2023, 15, 778. [Google Scholar] [CrossRef]
- Sirisha, U.; Praveen, S.P.; Srinivasu, P.N.; Barsocchi, P.; Bhoi, A.K. Statistical analysis of design aspects of various YOLO-based deep learning models for object detection. Int. J. Comput. Intell. Syst. 2023, 16, 126. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.K.; Girshick, R.B.; Farhadi, A. You Only Look Once: Unified, real-time object detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.E.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.B.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar] [CrossRef]
- Shen, Y.Y.; Liu, D.; Chen, J.Y.; Wang, Z.P.; Wang, Z.; Zhang, Q.L. On-board multi-class geospatial object detection based on convolutional neural network for High Resolution Remote Sensing Images. Remote Sens. 2023, 15, 3963. [Google Scholar] [CrossRef]
- Chen, Y.L.; Xu, H.L.; Zhang, X.J.; Gao, P.; Xu, Z.G.; Huang, X.B. An object detection method for bayberry trees based on an improved YOLO algorithm. Int. J. Digit. Earth 2023, 16, 781–805. [Google Scholar] [CrossRef]
- Wang, X.W.; Zhao, Q.Z.; Jiang, P.; Zheng, Y.C.; Yuan, L.M.Z.; Yuan, P.L. LDS-YOLO: A lightweight small object detection method for dead trees from shelter forest. Comput. Electron. Agric. 2022, 198, 107035. [Google Scholar] [CrossRef]
- Jintasuttisak, T.; Edirisinghe, E.; Elbattay, A. Deep neural network based date palm tree detection in drone imagery. Comput. Electron. Agric. 2022, 192, 106560. [Google Scholar] [CrossRef]
- Puliti, S.; Astrup, R. Automatic detection of snow breakage at single tree level using YOLOv5 applied to UAV imagery. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102946. [Google Scholar] [CrossRef]
- Dong, C.; Cai, C.Y.; Chen, S.; Xu, H.; Yang, L.B.; Ji, J.Y.; Huang, S.Q.; Hung, I.K.; Weng, Y.H.; Lou, X.W. Crown width extraction of Metasequoia glyptostroboides using improved YOLOv7 based on UAV images. Drones 2023, 7, 336. [Google Scholar] [CrossRef]
- YOLO v8. Available online: https://github.com/ultralytics/ultralytics (accessed on 25 March 2023).
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual tree-crown detection in RGB imagery using semi-supervised deep learning neural networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Perez, M.I.; Karelovic, B.; Molina, R.; Saavedra, R.; Cerulo, P.; Cabrera, G. Precision silviculture: Use of UAVs and comparison of deep learning models for the identification and segmentation of tree crowns in pine crops. Int. J. Digit. Earth 2022, 15, 2223–2238. [Google Scholar] [CrossRef]
- Choi, K.; Lim, W.; Chang, B.; Jeong, J.; Kim, I.; Park, C.R.; Ko, D.W. An automatic approach for tree species detection and profile estimation of urban street trees using deep learning and Google street view images. ISPRS J. Photogramm. Remote Sens. 2022, 190, 165–180. [Google Scholar] [CrossRef]
- Zhao, H.T.; Morgenroth, J.; Pearse, G.; Schindler, J. A systematic review of individual tree crown detection and delineation with convolutional neural networks (CNN). Curr. For. Rep. 2023, 9, 149–170. [Google Scholar] [CrossRef]
- Plesoianu, A.I.; Stupariu, M.S.; Sandric, I.; Patru-Stupariu, I.; Dragut, L. Individual tree-crown detection and species Classification in very high-resolution remote sensing imagery using a deep learning ensemble model. Remote Sens. 2020, 12, 2426. [Google Scholar] [CrossRef]
- Zhao, H.W.; Zhong, Y.F.; Wang, X.Y.; Hu, X.; Luo, C.; Boitt, M.; Piiroinen, R.; Zhang, L.P.; Heiskanen, J.; Pellikka, P. Mapping the distribution of invasive tree species using deep one-class classification in the tropical montane landscape of Kenya. ISPRS J. Photogramm. Remote Sens. 2022, 187, 328–344. [Google Scholar] [CrossRef]
- Zhong, H.; Lin, W.S.; Liu, H.R.; Ma, N.; Liu, K.K.; Cao, R.Z.; Wang, T.T.; Ren, Z.Z. Identification of tree species based on the fusion of UAV hyperspectral image and LiDAR data in a coniferous and broad-leaved mixed forest in Northeast China. Front. Plant Sci. 2022, 13, 964769. [Google Scholar] [CrossRef]
- Bai, Y.L.; Zhou, M.; Zhang, W.; Zhou, B.W.; Mei, T. Augmentation pathways network for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 10580–10587. [Google Scholar] [CrossRef]
- Guo, J.M.; Lou, H.T.; Chen, H.A.; Liu, H.Y.; Gu, J.S.; Bi, L.Y.; Duan, X.H. A new detection algorithm for alien intrusion on highway. Sci. Rep. 2023, 13, 10667. [Google Scholar] [CrossRef]
- Hazirbas, C.; Ma, L.; Domokos, C.; Cremers, D. FuseNet: Incorporating depth into semantic segmentation via fusion-based CNN architecture. In Proceedings of the Computer Vision—ACCV 2016, Taipei, Taiwan, 20–24 November 2016; pp. 213–228. [Google Scholar] [CrossRef]
- Sun, Y.M.; Cao, B.; Zhu, P.F.; Hu, Q.H. Drone-based RGB-Infrared cross-modality vehicle detection via uncertainty-aware learning. IEEE Trans. Circuits Syst. Video Technol. 2022, 32, 6700–6713. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the Computer Vision—ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar] [CrossRef]
- Ma, N.; Zhang, X.; Zheng, H.-T.; Sun, J. ShuffleNet V2: Practical Guidelines for Efficient CNN Architecture Design. In Proceedings of the Computer Vision—ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 122–138. [Google Scholar] [CrossRef]
- Wang, C.; He, W.; Nie, Y.; Guo, J.; Liu, C.; Han, K.; Wang, Y. Gold-YOLO: Efficient object detector via gather-and-distribute mechanism. arXiv 2023. [Google Scholar] [CrossRef]
- Niu, Y.; Cheng, W.; Shi, C.; Fan, S. YOLOv8-CGRNet: A lightweight object detection network leveraging context guidance and deep residual learning. Electronics 2024, 13, 43. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, G.; Ma, S.; Wang, Z.; Liu, H.; Gu, S. Potted phalaenopsis grading: Precise bloom and bud counting with the PA-YOLO algorithm and multiviewpoint imaging. Agronomy 2024, 14, 115. [Google Scholar] [CrossRef]
- Liu, B.; Wang, H.; Cao, Z.; Wang, Y.; Tao, L.; Yang, J.; Zhang, K. PRC-Light YOLO: An efficient lightweight model for fabric defect detection. Appl. Sci. 2024, 14, 938. [Google Scholar] [CrossRef]
- Wang, S.; Yan, B.; Xu, X.; Wang, W.; Peng, J.; Zhang, Y.; Wei, X.; Hu, W. Automated identification and localization of rail internal defects based on object detection networks. Appl. Sci. 2024, 14, 805. [Google Scholar] [CrossRef]
- Talaat, F.M.; ZainEldin, H. An improved fire detection approach based on YOLO-v8 for smart cities. Neural Comput. Appl. 2023, 35, 20939–20954. [Google Scholar] [CrossRef]
- Elmessery, W.M.; Gutiérrez, J.; Abd El-Wahhab, G.G.; Elkhaiat, I.A.; El-Soaly, I.S.; Alhag, S.K.; Al-Shuraym, L.A.; Akela, M.A.; Moghanm, F.S.; Abdelshafie, M.F. YOLO-based model for automatic detection of broiler pathological phenomena through visual and thermal images in intensive poultry houses. Agriculture 2023, 13, 1527. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Fricker, G.A.; Ventura, J.D.; Wolf, J.A.; North, M.P.; Davis, F.W.; Franklin, J. A convolutional neural network classifier identifies tree species in mixed-conifer forest from hyperspectral imagery. Remote Sens. 2019, 11, 2326. [Google Scholar] [CrossRef]
- Fu, Y.C.; Fan, J.F.; Xing, S.Y.; Wang, Z.; Jing, F.S.; Tan, M. Image segmentation of cabin assembly scene based on improved RGB-D Mask R-CNN. IEEE Trans. Instrum. Meas. 2022, 71, 5001512. [Google Scholar] [CrossRef]
- Xu, S.; Wang, R.; Shi, W.; Wang, X. Classification of tree species in transmission line corridors based on YOLO v7. Forests 2024, 15, 61. [Google Scholar] [CrossRef]


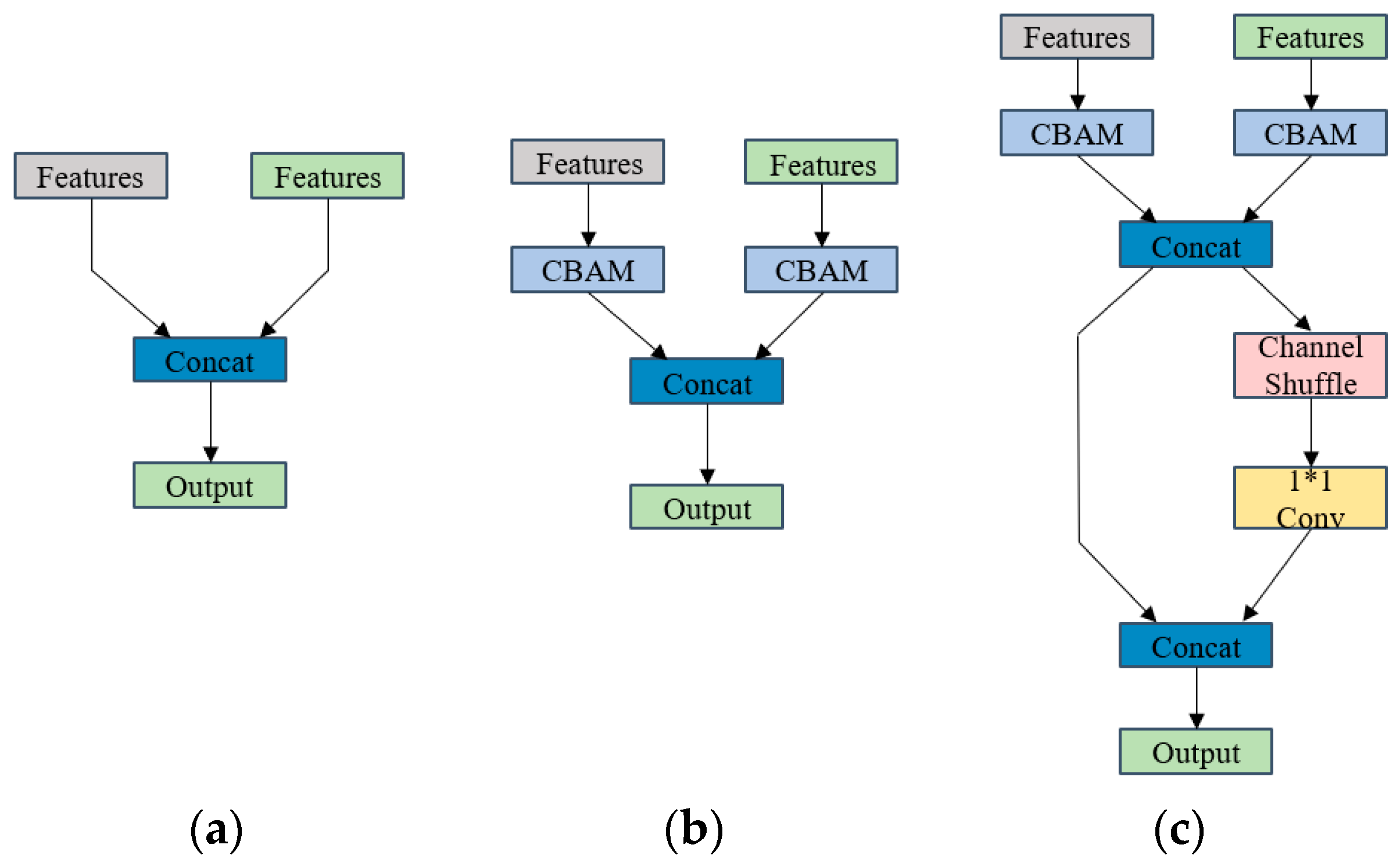


| Models | p | R | F1-Scores | mAP@50 |
|---|---|---|---|---|
| Retina Net | 0.623 | 0.534 | 0.575 | 55.1 |
| SSD | 0.645 | 0.568 | 0.604 | 61.2 |
| Faster R-CNN | 0.672 | 0.647 | 0.659 | 67.8 |
| YOLO v5x | 0.718 | 0.701 | 0.709 | 72.8 |
| YOLO v8x | 0.742 | 0.722 | 0.732 | 75.3 |
| Data | p | R | F1-Scores | mAP@50 |
|---|---|---|---|---|
| RGB | 0.742 | 0.722 | 0.732 | 75.3 |
| RG-D | 0.728 | 0.710 | 0.719 | 74.8 |
| D-GB | 0.741 | 0.729 | 0.735 | 75.5 |
| R-D-B | 0.741 | 0.721 | 0.732 | 75.2 |
| PCA-D | 0.747 | 0.739 | 0.743 | 76.2 |
| Data | Neck | Fusion Module | BP | PK | JM | LG | FM | PA | UP | OT | mAP@50 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RGB | PAN-FPN | - | 69.2 | 88.1 | 71.4 | 92.2 | 85.3 | 87 | 69.9 | 39.8 | 75.3 |
| RGB | GD | - | 69.8 | 88.4 | 72.5 | 92.6 | 86 | 87.7 | 70.9 | 40.2 | 76.0 |
| RGB + CHM | PAN-FPN | (a) | 73.7 | 91 | 75.7 | 93.5 | 85.5 | 89.2 | 75.7 | 46.3 | 78.8 |
| RGB + CHM | PAN-FPN | (b) | 75 | 91.3 | 76.1 | 93.7 | 86.3 | 89.7 | 77.4 | 48.7 | 79.2 |
| RGB + CHM | PAN-FPN | (c) | 75.7 | 91.4 | 76.5 | 93.7 | 87 | 90.1 | 78.7 | 50.8 | 80.3 |
| RGB + CHM | GD | (c) | 76.1 | 91.6 | 76.7 | 93.9 | 88.6 | 90.5 | 79.8 | 52.6 | 81.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, H.; Zhang, Z.; Liu, H.; Wu, J.; Lin, W. Individual Tree Species Identification for Complex Coniferous and Broad-Leaved Mixed Forests Based on Deep Learning Combined with UAV LiDAR Data and RGB Images. Forests 2024, 15, 293. https://doi.org/10.3390/f15020293
Zhong H, Zhang Z, Liu H, Wu J, Lin W. Individual Tree Species Identification for Complex Coniferous and Broad-Leaved Mixed Forests Based on Deep Learning Combined with UAV LiDAR Data and RGB Images. Forests. 2024; 15(2):293. https://doi.org/10.3390/f15020293
Chicago/Turabian StyleZhong, Hao, Zheyu Zhang, Haoran Liu, Jinzhuo Wu, and Wenshu Lin. 2024. "Individual Tree Species Identification for Complex Coniferous and Broad-Leaved Mixed Forests Based on Deep Learning Combined with UAV LiDAR Data and RGB Images" Forests 15, no. 2: 293. https://doi.org/10.3390/f15020293
APA StyleZhong, H., Zhang, Z., Liu, H., Wu, J., & Lin, W. (2024). Individual Tree Species Identification for Complex Coniferous and Broad-Leaved Mixed Forests Based on Deep Learning Combined with UAV LiDAR Data and RGB Images. Forests, 15(2), 293. https://doi.org/10.3390/f15020293






