CsAFS2 Gene from the Tea Plant Intercropped with Chinese Chestnut Plays an Important Role in Insect Resistance and Cold Resistance
Abstract
:1. Introduction
2. Results
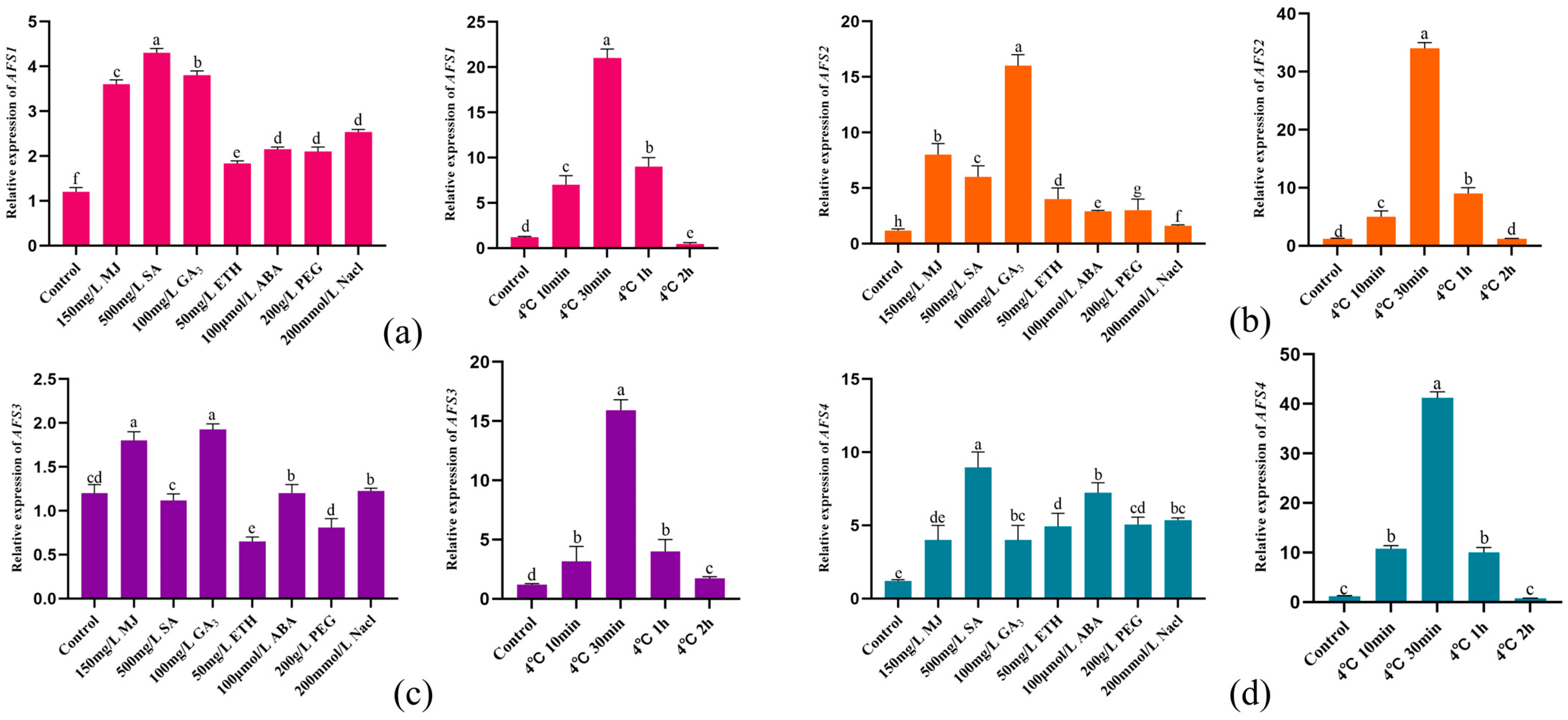
2.1. qRT-PCR Expression Analysis of AFS1, AFS2, AFS3, and AFS4
2.2. Gene Cloning and Sequence Analysis
2.3. Genetic Transformation in Tobacco
2.4. Generation and Screening of Transgenic Tobacco Lines
2.5. Phenotype Observation of the CsAFS2 Transgenic Lines
2.6. Analysis of Cold Resistance of CsAFS2-Overexpressing Tobacco Plants
2.7. Insect Resistance Analysis of CsAFS2-Overexpressing Tobacco Plants
2.8. The CsAFS2 Gene Promoter Analysis
3. Discussion
3.1. Effects of Trichomes on Plant Stress Resistance
3.2. The Combination Analysis of the CsAFS2 Gene and Transcription Factors
3.3. Regulation of CsAFS2 Gene by Hormone Crosstalks
3.4. CsAFS2 Might Enhance Insects and Cold Resistance in Plants by Releasing α-Farnesene
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Expression of AFS1, AFS2, AFS3, and AFS4 Genes under Different Plant Growth Regulators and Low Temperature
4.3. The CsAFS2 Gene Cloning
4.4. The CsAFS2 Gene Promoter Cloning and Sequence Analysis
4.5. Construction of Overexpression Vectors, Genetic Transformation and Identification of Transgenic Lines
4.6. Observations on the Phenotype of Transgenic Tobacco
4.7. Low-Temperature Stress in Transgenic Tobacco
4.8. Aphid Feeding Stress in Transgenic Tobacco
4.9. Determination of Physiological and Biochemical Indices of Transgenic Tobacco
4.10. Expression Analysis of Stress-Related Genes in Transgenic Tobacco
4.11. Statistical Analysis and Graphing of Data
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, X.; Zeng, L.; Liao, Y.; Li, J.; Tang, J.; Yang, Z. Formation of α-farnesene in tea (Camellia sinensis) leaves induced by herbivore-derived wounding and Its effect on neighboring tea plants. Int. J. Mol. Sci. 2019, 20, 4151. [Google Scholar] [CrossRef]
- Dong, F.; Yang, Z.; Baldermann, S.; Sato, Y.; Asai, T.; Watanabe, N. Herbivore-induced volatiles from tea (Camellia sinensis) plants and their involvement in intraplant communication and changes in endogenous nonvolatile metabolites. Agric. Food Chem. 2011, 29, 5–13. [Google Scholar] [CrossRef]
- Liao, Y.; Yu, Z.; Liu, X.; Zeng, L.; Cheng, S.; Li, J.; Tang, J.; Yang, Z. Effect of major tea insect attack on formation of quality-related nonvolatile specialized metabolites in tea (Camellia sinensis) leaves. J. Agric. Food Chem. 2019, 67, 6716–6724. [Google Scholar] [CrossRef]
- Lin, J.; Wang, D.; Chen, X.; Köllner, T.G.; Mazarei, M.; Guo, H.; Pantalone, V.R.; Arelli, P.; Stewart, C.N.; Wang, N.; et al. An (E,E)-α-farnesene synthase gene of soybean has a role in defence against nematodes and is involved in synthesizing insect-induced volatiles. Plant Biotechnol. J. 2017, 15, 510–519. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, H.; Zhang, M.; Liu, S.; Hao, Y.; Zhang, Y. MdMYC2 and MdERF3 positively co-regulate α-farnesene biosynthesis in apple. Front. Plant Sci. 2020, 11, 512844. [Google Scholar] [CrossRef]
- Wu, S.; Yang, Y.; Chen, J.; Li, J.; Jian, G.; Yang, J.; Mao, K.; Zeng, L.; Gu, D. Histone deacetylase CsHDA6 mediates the regulated formation of the anti-insect metabolite α-farnesene in tea (Camellia sinensis). Plant Sci. 2023, 326, 111501. [Google Scholar] [CrossRef]
- Souleyre, E.J.F.; Bowen, J.K.; Matich, A.J.; Tomes, S.; Chen, X.; Hunt, M.B.; Wang, M.Y.; Ileperuma, N.R.; Richards, K.; Rowan, D.D.; et al. Genetic control of α-farnesene production in apple fruit and its role in fungal pathogenesis. Plant J. 2019, 100, 1148–1162. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; da Silva, J.A.T.; Zhang, T.; Xiong, Y.; Li, Y.; Yuan, Y.; Pan, X.; Ma, G. Characterization of sandalwood (E,E)-α-farnesene synthase whose overexpression enhances cold tolerance through jasmonic acid biosynthesis and signaling in Arabidopsis. Planta 2023, 258, 1–21. [Google Scholar] [CrossRef]
- Pokharel, S.S.; Yu, H.; Fang, W.; Parajulee, M.N.; Chen, F. Intercropping cover crops for a vital ecosystem service: A review of the biocontrol of insect pests in tea agroecosystems. Plants 2023, 12, 2361. [Google Scholar] [CrossRef]
- Wang, T.; Duan, Y.; Liu, G.; Shang, X.; Liu, L.; Zhang, K.; Li, J.; Zou, Z.; Zhu, X.; Fang, W. Tea plantation intercropping green manure enhances soil functional microbial abundance and multifunctionality resistance to drying-rewetting cycles. Sci. Total Environ. 2022, 810, 151282. [Google Scholar] [CrossRef]
- Wu, T.; Jiang, Y.; Li, M.; Pu, D.; Shi, M.; Lan, Z. RNA-seq analysis reveals the potential mechanism of improved viability and product quality of tea plants through intercropping with Chinese chestnut. Plant Growth Regul. 2022, 96, 177–193. [Google Scholar] [CrossRef]
- Meng, Y.; Lyu, X.; Liu, J.; Gao, W.; Ma, Y.; Liao, N.; Li, Z.; Bo, Y.; Hu, Z.; Yang, J.; et al. Structural variation of GL1 gene determines the trichome formation in Brassica juncea. Theor. Appl. Genet. 2023, 136, 75. [Google Scholar] [CrossRef] [PubMed]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Bak, S. Plant defense against insect herbivores. Int. J. Mol. Sci. 2013, 14, 10242. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yan, X.; Zhang, H.; Meng, Y.; Pan, Y.; Cui, H. NtCycB2 negatively regulates tobacco glandular trichome formation, exudate accumulation, and aphid resistance. Plant Mol. Biol. 2022, 108, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Spyropoulou, E.A.; Haring, M.A.; Schuurink, R.C. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 2014, 15, 402. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, T.; Zhong, J.; Ba, T.; Xu, T.; Zhang, Q.; Sun, M. Identification of the volatile compounds and observation of the glandular trichomes in Opisthopappus taihangensis and four species of chrysanthemum. Plants 2020, 9, 855. [Google Scholar] [CrossRef]
- Cao, H.; Li, J.; Ye, Y.; Lin, H.; Hao, Z.; Ye, N.; Yue, C. Integrative transcriptomic and metabolic analyses provide insights into the role of trichomes in tea plant (Camellia sinensis). Biomolecules 2020, 10, 311. [Google Scholar] [CrossRef]
- Zeng, L.; Liao, Y.; Li, J.; Zhou, Y.; Tang, J.; Dong, F.; Yang, Z. α-Farnesene and ocimene induce metabolite changes by volatile signaling in neighboring tea (Camellia sinensis) plants. Plant Sci. 2017, 264, 29–36. [Google Scholar] [CrossRef]
- Chen, X.; Wang, M.Y.; Deng, C.H.; Beatson, R.A.; Templeton, K.R.; Atkinson, R.G.; Nieuwenhuizen, N.J. The hops (Humulus lupulus) genome contains a mid-sized terpene synthase family that shows wide functional and allelic diversity. BMC Plant Biol. 2023, 23, 280. [Google Scholar] [CrossRef]
- Xu, J.; van Herwijnen, Z.O.; Dräger, D.B.; Sui, C.; Haring, M.A.; Schuurink, R.C. SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef]
- Hua, B.; Chang, J.; Wu, M.; Xu, Z.; Zhang, F.; Yang, M.; Xu, H.; Wang, L.; Chen, X.; Wu, S. Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato. Plant Biotechnol. J. 2020, 19, 375–393. [Google Scholar] [CrossRef]
- Zhao, Q.; Xiang, X.; Liu, D.; Yang, A.; Wang, Y. Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front. Plant Sci. 2018, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.A.; Wang, Q.; Dhar, N.; Kumar, N.; Venkatesh, P.N.; Rajan, C.; Panicker, D.; Sridhar, V.; Mao, H.; Sarojam, R. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 2017, 15, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; He, M.; Wang, Z.; Xu, J. Integrative analysis of terpenoid profiles and hormones from fruits of red-flesh citrus mutants and their wild types. Molecules 2019, 24, 3456. [Google Scholar] [CrossRef] [PubMed]
- Body, M.J.A.; Neer, W.C.; Vore, C.; Lin, C.-H.; Vu, D.C.; Schultz, J.C.; Cocroft, R.B.; Appel, H.M. Caterpillar chewing vibrations cause changes in plant hormones and volatile emissions in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 810. [Google Scholar] [CrossRef]
- Jing, T.; Qian, X.; Du, W.; Gao, T.; Li, D.; Guo, D.; He, F.; Yu, G.; Li, S.; Schwab, W.; et al. Herbivore-induced volatiles influence moth preference by increasing the β-Ocimene emission of neighbouring tea plants. Plant Cell Environ. 2021, 44, 3667–3680. [Google Scholar] [CrossRef]
- Jia, D.; Gao, S.; Duan, P.; Chen, J.; Tian, F.; Yu, X. Metabolic engineering of (E)-β-farnesene synthase genes for aphid-resistant genetically modified plants. Sheng Wu Gong Cheng Xue Bao 2018, 34, 12–23. [Google Scholar] [CrossRef]
- Warneys, R.; Gaucher, M.; Robert, P.; Aligon, S.; Anton, S.; Aubourg, S.; Barthes, N.; Braud, F.; Cournol, R.; Gadenne, C.; et al. Acibenzolar-S-Methyl reprograms apple transcriptome toward resistance to rosy apple Aphid. Front. Plant Sci. 2018, 9, 1795. [Google Scholar] [CrossRef]
- Song, J.; Wu, H.; He, F.; Qu, J.; Wang, Y.; Li, C.; Liu, J.-H. Citrus sinensis CBF1 functions in cold tolerance by modulating putrescine biosynthesis through regulation of arginine decarboxylase. Plant Cell Physiol. 2021, 63, 19–29. [Google Scholar] [CrossRef]
- Perez-Garcia, P.; Pucciariello, O.; Sanchez-Corrionero, A.; Cabrera, J.; del Barrio, C.; del Pozo, J.C.; Perales, M.; Wabnik, K.; Moreno-Risueno, M.A. The cold-induced factor CBF3 mediates root stem cell activity, regeneration, and developmental responses to cold. Plant Commun. 2023, 4, 100737. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Dai, Y.; He, Y.; Li, C.; Mao, P.; Ma, X. The transcription factors of tall fescue in response to temperature stress. Plant Biol. 2021, 23, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, X.; Zhou, J.; Dong, Z.; Yu, H.; Tang, Q.; Yuan, L.; Peng, S.; Zhong, X.; He, Y. Selection and verification of standardized reference genes of Angelica dahurica under various abiotic stresses by real-time quantitative PCR. Genes 2024, 15, 79. [Google Scholar] [CrossRef] [PubMed]
- Vaghela, B.; Vashi, R.; Rajput, K.; Joshi, R. Plant chitinases and their role in plant defense: A comprehensive review. Enzym. Microb. Technol. 2022, 159, 110055. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Cha, J.-H.; Wang, H.-C.; Ye, P.; Chen, B.; Chen, M.; Yang, W.-H.; Yan, X. An undefined cystatin CsCPI1 from tea plant Camellia sinensis harbors antithrombotic activity. Biomed. Pharmacother. 2023, 159, 114285. [Google Scholar] [CrossRef]
- Schäfer, M.; Meza-Canales, I.D.; Navarro-Quezada, A.; Brütting, C.; Vanková, R.; Baldwin, I.T.; Meldau, S. Cytokinin levels and signaling respond to wounding and the perception of herbivore elicitors in Nicotiana attenuata. J. Integr. Plant Biol. 2015, 57, 198–212. [Google Scholar] [CrossRef]
- Walley, J.W.; Kelley, D.R.; Savchenko, T.; Dehesh, K. Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal. Behav. 2010, 5, 802–805. [Google Scholar] [CrossRef]









| Possible Binding Transcription Factors | Core Sequence | Possible Functions | Name of cis-Element | Number |
|---|---|---|---|---|
| WRKY | TGAC | TGAC core containing W-box, transcriptional repressor of gibberellin signaling pathway | WRKY71OS | 7 |
| WRKY, ERF | TGACY | Involved in activation of ERF3 gene by wounding | WBOXNTERF3 | 2 |
| WRKY | CTGACY | Involved in elicitor-responsive transcription of defense genes | WBOXNTCHN48 | 2 |
| WRKY | TTGAC | Pathogen- and SA-responsive element | WBOXATNPR1 | 2 |
| WRKY | TTTTTTCC | GA induction | PYRIMIDNEBOXHVEPB1 | 1 |
| MYB | CNGTTR | Regulation of genes involved in water stress response | MYB-CORE | 5 |
| MYB | TAACAAAA | GA-regulated transcription factor | MYBGAHV | 5 |
| MYB | TAACAGA | GA-responsive element | GARE1OSRER1 | 4 |
| MYB | MACCWAMC | Regulate phenylpropanoid and lignin biosynthesis | MYBPLANT | 3 |
| MYB | WAACCA | ABA-responsive element | MYB1AT | 4 |
| ERF | GCCGCC | Ethylene responsive element | GCC-CORE | 2 |
| ERF | WTTSSCSS | Secondary transcription factor that triggers ethylene signaling | ERFAT | 4 |
| bZIP | AACGTG | Pathogen- and JA-responsive element | T/GBOXATPIN2 | 3 |
| PBF | ACACNNG | ABA-responsive element | DPBFCOREDCDC3 | 4 |
| bHLH | CCGAAA | Cold-responsive elements | LTRE1HVBLT49 | 2 |
| MYC | CANNTG | MYCCONSENSUSAT | MYCCONSENSUSAT | 8 |
| Possible Binding Transcription Factors | Gene Id | Description | E-Value | Log2FC | |
|---|---|---|---|---|---|
| Pure Tea Plantation | Intercropped Tea Plantation | ||||
| WRKY | LOC114289849 | WRKY1 | 4.00 × 10−110 | 9.52 ± 0.33 | 12.08 ± 0.51 |
| LOC114315408 | WRKY105 | 4.00 × 10−52 | 14.68 ± 0.26 | 19.65 ± 0.41 | |
| LOC114314481 | WRKY6 | 7.00 × 10−102 | 8.21 ± 0.17 | 10.43 ± 0.71 | |
| LOC114299830 | WRKY8 | 5.00 × 10−99 | 3.38 ± 0.08 | 5.79 ± 0.13 | |
| LOC114256467 | WRKY24 | 2.00 × 10−142 | 14.36 ± 0.63 | 15.01 ± 0.86 | |
| LOC114284484 | WRKY45 | 9.00 × 10−38 | 0 ± 0.00 | 0.06 ± 0.01 | |
| LOC114287654 | WRKY23 | 1.00 × 10−51 | 0.48 ± 0.04 | 0.18 ± 0.02 | |
| LOC114299833 | WRKY38 | 3.00 × 10−53 | 2.23 ± 0.07 | 1.08 ± 0.09 | |
| LOC114275007 | WRKY10 | 6.00 × 10−13 | 5.25 ± 0.11 | 5.27 ± 0.24 | |
| LOC114259259 | WRKY70 | 6.00 × 10−30 | 7.22 ± 0.37 | 6.84 ± 0.43 | |
| MYB | LOC114284747 | MYB36 | 1.00 × 10−137 | 0.04 ± 0.01 | 0 ± 0.00 |
| LOC114271197 | MYB44 | 5.00 × 10−25 | 3.57 ± 0.59 | 5.76 ± 0.64 | |
| LOC114277061 | MYB88 | 4.00 × 10−120 | 5.17 ± 0.22 | 5.07 ± 0.18 | |
| LOC114262164 | MYB105 | 3.00 × 10−36 | 0.07 ± 0.02 | 0 ± 0.00 | |
| LOC114308355 | MYB4 | 2.00 × 10−54 | 16.89 ± 1.22 | 16.38 ± 1.37 | |
| LOC114262089 | MYB16 | 3.00 × 10−117 | 38.28 ± 1.45 | 40.24 ± 1.47 | |
| LOC114322988 | MYB86 | 3.00 × 10−89 | 3.88 ± 0.76 | 3.31 ± 0.45 | |
| LOC114295878 | MYB101 | 1.00 × 10−67 | 0.04 ± 0.00 | 0 ± 0.00 | |
| LOC114277090 | MYB5 | 8.00 × 10−114 | 17.56 ± 2.37 | 16.64 ± 1.56 | |
| LOC114277091 | MYB10 | 3.00 × 10−36 | 6.03 ± 0.84 | 3.83 ± 0.77 | |
| LOC114316900 | MYB35 | 5.00 × 10−110 | 0.04 ± 0.00 | 0.05 ± 0.00 | |
| LOC114309279 | MYB61 | 1.00 × 10−121 | 5.21 ± 1.24 | 5.52 ± 1.57 | |
| LOC114292749 | MYB2 | 1.00 × 10−114 | 0.05 ± 0.02 | 0 ± 0.00 | |
| LOC114285525 | MYB52 | 2.00 × 10−100 | 0.18 ± 0.07 | 0.63 ± 0.14 | |
| LOC114297472 | MYB15 | 3.00 × 10−83 | 1.78 ± 0.66 | 2.08 ± 1.57 | |
| LOC114279142 | MYB39 | 6.00 × 10−81 | 0.52 ± 0.13 | 0.64 ± 0.37 | |
| LOC114276260 | MYB98 | 2.00 × 10−67 | 0.34 ± 0.08 | 0.19 ± 0.04 | |
| LOC114268476 | MYB110 | 6.00 × 10−49 | 0.06 ± 0.02 | 0.11 ± 0.00 | |
| LOC114281490 | MYB111 | 1.00 × 10−53 | 3.71 ± 0.40 | 3.14 ± 0.32 | |
| ERF | LOC114312023 | ERF2 | 2.00 × 10−68 | 1.19 ± 0.06 | 2.44 ± 0.09 |
| LOC114319131 | ERF16 | 6.00 × 10−43 | 2.34 ± 0.04 | 2.69 ± 0.03 | |
| LOC114260629 | ERF18 | 1.00 × 10−57 | 0.88 ± 0.22 | 0.96 ± 0.35 | |
| LOC114264712 | ERF61 | 2.00 × 10−54 | 4.88 ± 0.14 | 7.69 ± 0.27 | |
| LOC114288203 | ERF109 | 9.00 × 10−53 | 1.41 ± 0.11 | 2.18 ± 0.17 | |
| LOC114257849 | ERF115 | 9.00 × 10−59 | 6.37 ± 0.28 | 11.76 ± 0.37 | |
| LOC114275420 | ERF3 | 1.00 × 10−38 | 2.55 ± 0.34 | 4.83 ± 0.95 | |
| LOC114268064 | ERF20 | 6.00 × 10−14 | 0.08 ± 0.00 | 0 ± 0.00 | |
| LOC114284734 | ERF118 | 1.00 × 10−8 | 22.04 ± 1.46 | 16.98 ± 1.87 | |
| LOC114312929 | ERF24 | 3.00 × 10−54 | 0.07 ± 0.00 | 0 ± 0.00 | |
| LOC114294084 | ERF26 | 2.00 × 10−19 | 2.11 ± 0.24 | 0.89 ± 0.16 | |
| LOC114277320 | ERF96 | 4.00 × 10−6 | 0.14 ± 0.00 | 0 ± 0.00 | |
| LOC114290124 | ERF114 | 3.00 × 10−44 | 0.99 ± 0.19 | 2.08 ± 0.42 | |
| LOC114300795 | ERF38 | 4.00 × 10−41 | 3.65 ± 0.79 | 4.38 ± 1.20 | |
| LOC114255784 | ERF34 | 3.00 × 10−27 | 15.04 ± 1.06 | 11.92 ± 1.11 | |
| MYC | LOC114283260 | MYC2 | 0 | 62.66 ± 2.35 | 50.51 ± 1.97 |
| bHLH | LOC114261778 | bHLH96 | 9.00 × 10−95 | 12.53 ± 1.09 | 19.05 ± 1.81 |
| LOC114318495 | bHLH162 | 2.00 × 10−28 | 3.66 ± 0.03 | 6.47 ± 1.54 | |
| LOC114280978 | bHLH123 | 8.00 × 10−91 | 1.94 ± 0.72 | 3.18 ± 0.94 | |
| LOC114320661 | bHLH96 | 9.00 × 10−95 | 47.71 ± 1.65 | 30.17 ± 1.87 | |
| LOC114263002 | bHLH74 | 2.00 × 10−172 | 17.44 ± 1.47 | 16.19 ± 1.75 | |
| LOC114289221 | bHLH120 | 5.00 × 10−31 | 0.69 ± 0.07 | 0.62 ± 0.00 | |
| LOC114288662 | bHLH110 | 4.00 × 10−78 | 6.09 ± 1.01 | 7.27 ± 1.25 | |
| LOC114281839 | bHLH111 | 7.00 × 10−81 | 0.48 ± 0.00 | 0.52 ± 0.08 | |
| bZIP | LOC114315402 | bZIP1 | 7.00 × 10−22 | 1.09 ± 0.27 | 1.84 ± 0.62 |
| LOC114279212 | bZIP2 | 4.00 × 10−93 | 3.59 ± 0.12 | 5.28 ± 0.31 | |
| LOC114307154 | bZIP11 | 0 | 23.78 ± 0.97 | 24.26 ± 0.84 | |
| Gene Name | GenBank No. | Forward Primer (5′–3′) | Reforward Primer (5′–3′) |
|---|---|---|---|
| NtCBF1 [29] | NP001312156 | GGATGAGGAGACGCTATTCTG | TGTGAACACTGAGGTGGAGG |
| NtCBF3 [30] | NP001312741 | TGTGAACACTGAGGTGGAGG | CCTCCTCGTCCATAAACAA |
| NtDREB2B [31] | EU727156 | CGGCCGCCCATCTGAGTC | AGGTGGAGGCAGCATTAGTC |
| NtCOR4 [32] | NW015826227.1 | TGTCATCGAAAAGCTTCACCGA | TGTCATCGAAAAGCTTCACCGA |
| EF1α [33] | NM001326165 | TGGTTGTGACTTTTGGTCCCA | ACAAACCCACGCTTGAGATCC |
| Gene Name | GenBank No. | Forward Primer (5′–3′) | Reforward Primer (5′–3′) |
|---|---|---|---|
| NtChiA [34] | P08252.2 | GGCCTTGTGGAAGAGCCATA | CCAAATCCAGGGAGGCGATT |
| NtCPI1 [35] | KF0S7988 | TCTGGAGTTCGGAAAGGTTGTT | CAGACCTTGGCTTCGTATGCT |
| NtTD [36] | AAG59585.1 | ACATGGGTCAAGTTAGGCGG | TATAGGGGTGGCAAATGGGC |
| NtCAF1 [37] | NP001312482.1 | ATCATCATCACGCGGTCGAA | TTTTGCTGAAAGCTGCCGAC |
| EF1α [33] | NM001326165 | TGGTTGTGACTTTTGGTCCCA | ACAAACCCACGCTTGAGATCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Dao, M.; Yang, Z.; Bai, Y.; Qin, Y.; Wu, T. CsAFS2 Gene from the Tea Plant Intercropped with Chinese Chestnut Plays an Important Role in Insect Resistance and Cold Resistance. Forests 2024, 15, 380. https://doi.org/10.3390/f15020380
Wang J, Dao M, Yang Z, Bai Y, Qin Y, Wu T. CsAFS2 Gene from the Tea Plant Intercropped with Chinese Chestnut Plays an Important Role in Insect Resistance and Cold Resistance. Forests. 2024; 15(2):380. https://doi.org/10.3390/f15020380
Chicago/Turabian StyleWang, Jianzhao, Mei Dao, Ziyun Yang, Yan Bai, Ying Qin, and Tian Wu. 2024. "CsAFS2 Gene from the Tea Plant Intercropped with Chinese Chestnut Plays an Important Role in Insect Resistance and Cold Resistance" Forests 15, no. 2: 380. https://doi.org/10.3390/f15020380
APA StyleWang, J., Dao, M., Yang, Z., Bai, Y., Qin, Y., & Wu, T. (2024). CsAFS2 Gene from the Tea Plant Intercropped with Chinese Chestnut Plays an Important Role in Insect Resistance and Cold Resistance. Forests, 15(2), 380. https://doi.org/10.3390/f15020380