Estimating the Aboveground Biomass of Robinia pseudoacacia Based on UAV LiDAR Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data Sources and Processing
2.2.1. Sample Survey
2.2.2. LiDAR Data Acquisition and Processing
2.2.3. Split Window Size
2.2.4. Individual Tree Extraction and Segmentation
2.2.5. Verification of Individual Tree Segmentation Results
3. Results
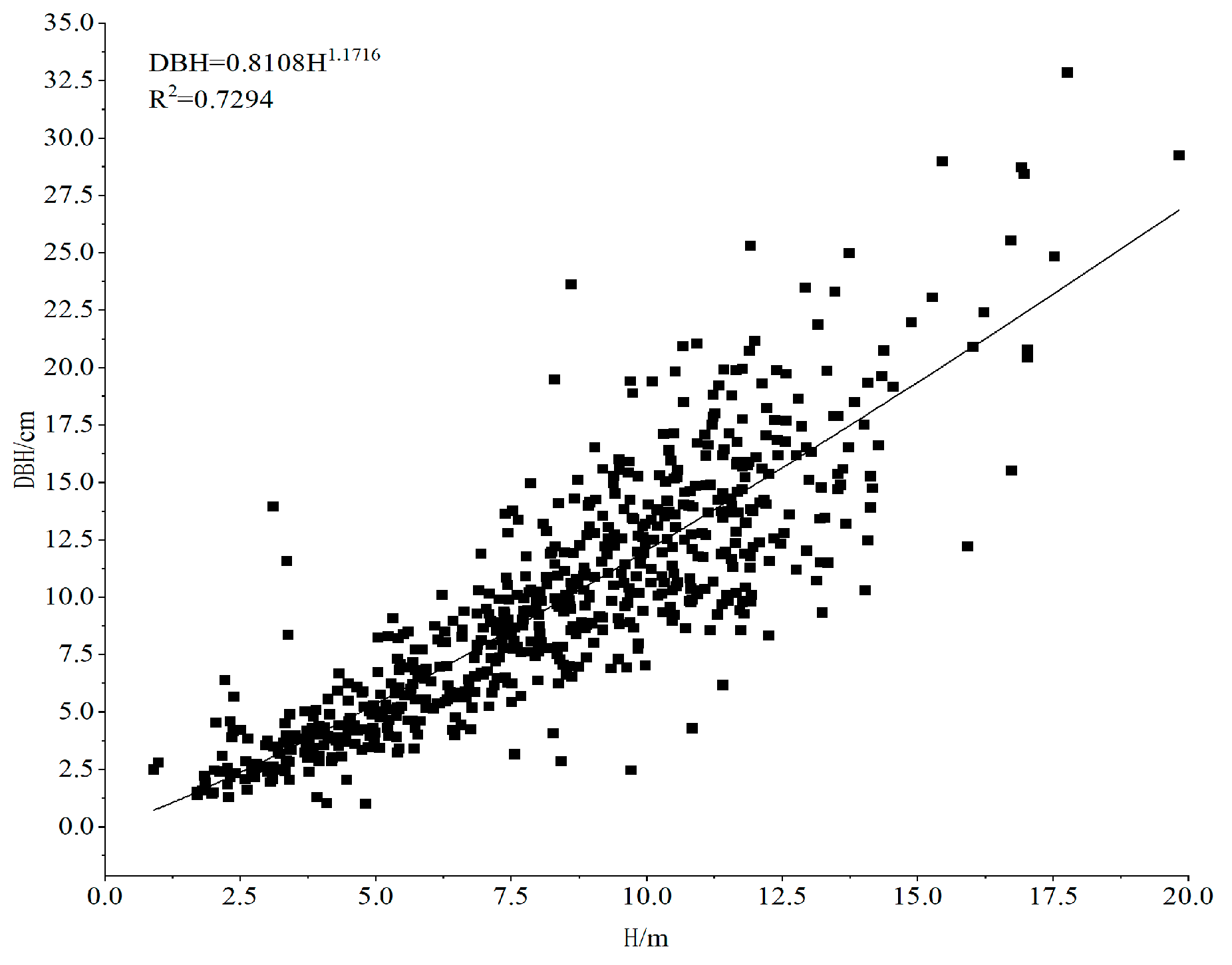
3.1. Height–Diameter–Biomass Models
3.2. Analysis of Segmentation Window Size and Individual Tree Recognition Accuracy
3.3. Results and Analysis of Extracted Tree Height
3.4. Results and Analysis of Extracted DBH
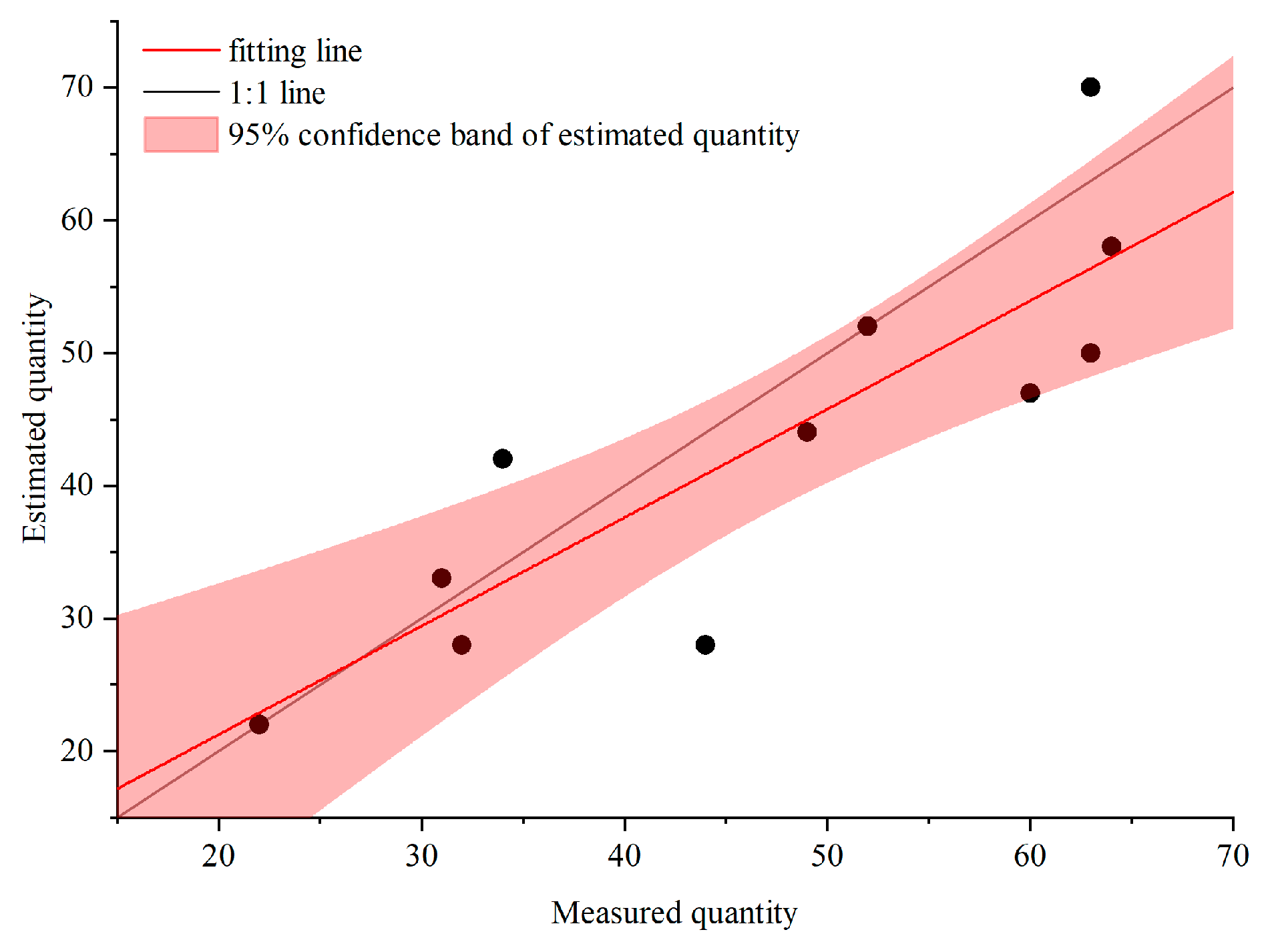
3.5. Estimation and Analysis of Biomass
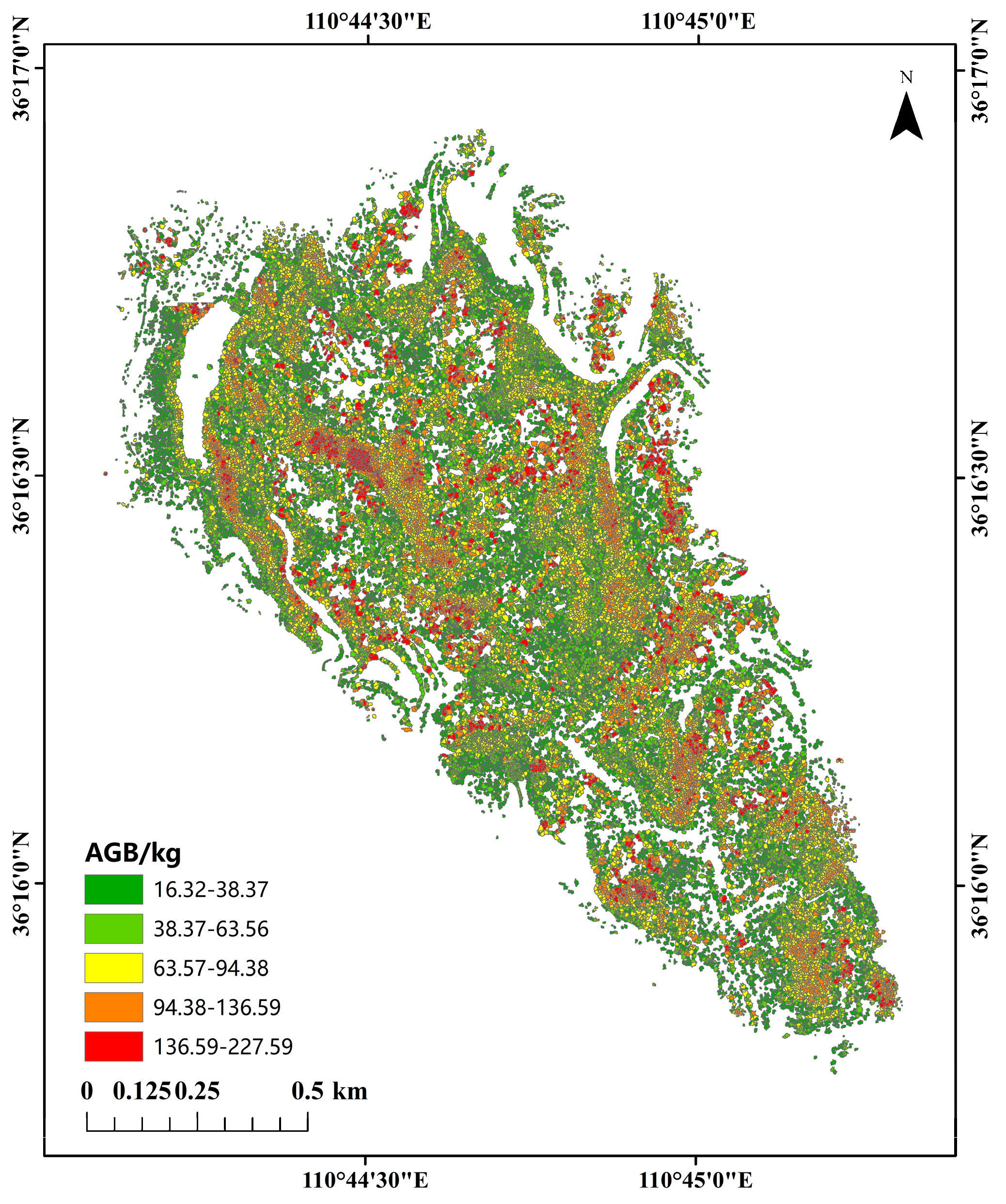
3.6. Biomass Estimation and Analysis in the Study Area
4. Discussion
4.1. Influencing Factors of Individual Tree Segmentation
4.2. Influencing Factors of Tree Height Extraction
4.3. Influencing Factors of DBH Extraction
4.4. Influencing Factors of Biomass Estimation Accuracy
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piao, S.; Fang, J.; He, J.; Xiao, Y. Grassland vegetation biomass and its spatial distribution pattern in China. Chin. J. Plant Ecol. 2004, 4, 491–498. [Google Scholar]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Kumar, L.; Mutanga, O. Remote sensing of above-ground biomass. Remote Sens. 2017, 9, 935. [Google Scholar] [CrossRef]
- Hudson, N.W. Soil Conservation; Scientific Publishers: Singapore, 2015. [Google Scholar]
- Troeh, F.R.; Hobbs, J.A.; Donahue, R.L. Soil and Water Conservation for Productivity and Environmental Protection; Prentice-Hall, Inc.: Upper Saddle River, NJ, USA, 1980. [Google Scholar]
- Kahilainen, A.; Puurtinen, M.; Kotiaho, J.S. Conservation implications of species–genetic diversity correlations. Glob. Ecol. Conserv. 2014, 2, 315–323. [Google Scholar] [CrossRef]
- Van Impe, W.F. Soil Improvement Techniques and Their Evolution; Taylor & Francis: Abingdon, UK, 1989. [Google Scholar]
- Dong, L.; Zhang, L.; Li, F. Developing two additive biomass equations for three coniferous plantation species in Northeast China. Forests 2016, 7, 136. [Google Scholar] [CrossRef]
- Bond-Lamberty, B.; Wang, C.; Gower, S. Aboveground and belowground biomass and sapwood area allometric equations for six boreal tree species of northern Manitoba. Can. J. For. Res. 2002, 32, 1441–1450. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Sarker, L.R.; Nichol, J.E. Improved forest biomass estimates using ALOS AVNIR-2 texture indices. Remote Sens. Environ. 2011, 115, 968–977. [Google Scholar] [CrossRef]
- Kachamba, D.J.; Ørka, H.O.; Gobakken, T.; Eid, T.; Mwase, W. Biomass estimation using 3D data from unmanned aerial vehicle imagery in a tropical woodland. Remote Sens. 2016, 8, 968. [Google Scholar] [CrossRef]
- Deng, S.; Katoh, M.; Guan, Q.; Yin, N.; Li, M. Estimating forest aboveground biomass by combining ALOS PALSAR and WorldView-2 data: A case study at Purple Mountain National Park, Nanjing, China. Remote Sens. 2014, 6, 7878–7910. [Google Scholar] [CrossRef]
- Návar, J. Measurement and assessment methods of forest aboveground biomass: A literature review and the challenges ahead. In Biomass; Momba, M., Bux, F., Eds.; InTech: Rijeka, Croatia, 2010; pp. 27–64. [Google Scholar]
- Englhart, S.; Keuck, V.; Siegert, F. Aboveground biomass retrieval in tropical forests—The potential of combined X-and L-band SAR data use. Remote Sens. Environ. 2011, 115, 1260–1271. [Google Scholar] [CrossRef]
- Sarker, M.L.R.; Nichol, J.; Ahmad, B.; Busu, I.; Rahman, A.A. Potential of texture measurements of two-date dual polarization PALSAR data for the improvement of forest biomass estimation. ISPRS J. Photogramm. Remote Sens. 2012, 69, 146–166. [Google Scholar] [CrossRef]
- Tsitsi, B. Remote sensing of aboveground forest biomass: A review. Trop. Ecol. 2016, 57, 125–132. [Google Scholar]
- Lefsky, M.A.; Cohen, W.B.; Harding, D.J.; Parker, G.G.; Acker, S.A.; Gower, S.T. Lidar remote sensing of above-ground biomass in three biomes. Glob. Ecol. Biogeogr. 2002, 11, 393–399. [Google Scholar] [CrossRef]
- Torre-Tojal, L.; Bastarrika, A.; Boyano, A.; Lopez-Guede, J.M.; Grana, M. Above-ground biomass estimation from LiDAR data using random forest algorithms. J. Comput. Sci. 2022, 58, 101517. [Google Scholar] [CrossRef]
- Meng, Y.; Dong, X.; Liu, W.; Lin, W. Modeling biomass of white birch (Betula platyphylla) in the Lesser Khingan Range of China based on terrestrial 3D laser scanning system. Nat. Resour. Model. 2020, 33, e12240. [Google Scholar] [CrossRef]
- Yuan, Z.; Gazol, A.; Wang, X.; Lin, F.; Ye, J.; Zhang, Z.; Suo, Y.; Kuang, X.; Wang, Y.; Jia, S. Pattern and dynamics of biomass stock in old growth forests: The role of habitat and tree size. Acta Oecol. 2016, 75, 15–23. [Google Scholar] [CrossRef]
- Li, W.; Niu, Z.; Gao, S.; Huang, N.; Chen, H. Correlating the horizontal and vertical distribution of lidar point clouds with components of biomass in a Picea crassifolia forest. Forests 2014, 5, 1910–1930. [Google Scholar] [CrossRef]
- Zhen, Z.; Yang, L.; Ma, Y.; Wei, Q.; Jin, H.I.; Zhao, Y. Upscaling aboveground biomass of larch (Larix olgensis Henry) plantations from field to satellite measurements: A comparison of individual tree-based and area-based approaches. GIScience Remote Sens. 2022, 59, 722–743. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, W.; Ji, Y.; Huang, J.; Huang, G.; Wang, L. Total and component forest aboveground biomass inversion via LiDAR-derived features and machine learning algorithms. Front. Plant Sci. 2023, 14, 1258521. [Google Scholar] [CrossRef]
- Nie, S.; Wang, C.; Zeng, H.; Xi, X.; Li, G. Above-ground biomass estimation using airborne discrete-return and full-waveform LiDAR data in a coniferous forest. Ecol. Indic. 2017, 78, 221–228. [Google Scholar] [CrossRef]
- Qin, H.; Wang, C.; Xi, X.; Tian, J.; Zhou, G. Estimation of coniferous forest aboveground biomass with aggregated airborne small-footprint LiDAR full-waveforms. Opt. Express 2017, 25, A851–A869. [Google Scholar] [CrossRef]
- He, Q. Estimation of coniferous forest above-ground biomass using LiDAR and SPOT-5 data. In Proceedings of the 2012 2nd International Conference on Remote Sensing, Environment and Transportation Engineering, Nanjing, China, 1–3 June 2012; pp. 1–4. [Google Scholar]
- Wang, X.; Liu, C.; Lv, G.; Xu, J.; Cui, G. Integrating multi-source remote sensing to assess forest aboveground biomass in the Khingan mountains of north-eastern China using machine-learning algorithms. Remote Sens. 2022, 14, 1039. [Google Scholar] [CrossRef]
- Qi, Z.; Li, S.; Pang, Y.; Du, L.; Zhang, H.; Li, Z. Monitoring Spatiotemporal Variation of Individual Tree Biomass Using Multitemporal LiDAR Data. Remote Sens. 2023, 15, 4768. [Google Scholar] [CrossRef]
- Zhou, L.; Li, X.; Zhang, B.; Xuan, J.; Gong, Y.; Tan, C.; Huang, H.; Du, H. Estimating 3D Green Volume and Aboveground Biomass of Urban Forest Trees by UAV-Lidar. Remote Sens. 2022, 14, 5211. [Google Scholar] [CrossRef]
- d’Oliveira, M.V.; Broadbent, E.N.; Oliveira, L.C.; Almeida, D.R.; Papa, D.A.; Ferreira, M.E.; Zambrano, A.M.A.; Silva, C.A.; Avino, F.S.; Prata, G.A. Aboveground biomass estimation in Amazonian tropical forests: A comparison of aircraft-and gatoreye UAV-borne LIDAR data in the Chico mendes extractive reserve in Acre, Brazil. Remote Sens. 2020, 12, 1754. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, J.; Wan, P.; Wang, H.; Xie, D.; Wang, X.; Yan, G. An easy-to-use airborne LiDAR data filtering method based on cloth simulation. Remote Sens. 2016, 8, 501. [Google Scholar] [CrossRef]
- Changsai, Z.; Zhengjun, L.; Shuwen, Y.; Zhiquan, Z. Applicability analysis of cloth simulation filtering algorithm based on LiDAR data. Laser Technol. 2018, 42, 410–416. [Google Scholar]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef]
- Yin, D.; Wang, L. Individual mangrove tree measurement using UAV-based LiDAR data: Possibilities and challenges. Remote Sens. Environ. 2019, 223, 34–49. [Google Scholar] [CrossRef]
- Roerdink, J.B.; Meijster, A. The watershed transform: Definitions, algorithms and parallelization strategies. Fundam. Informaticae 2000, 41, 187–228. [Google Scholar] [CrossRef]
- Najman, L.; Schmitt, M. Watershed of a continuous function. Signal Process. 1994, 38, 99–112. [Google Scholar] [CrossRef]
- Goutte, C.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In Proceedings of the European Conference on Information Retrieval, Santiago de Compostela, Spain, 21–23 March 2005; pp. 345–359. [Google Scholar]
- Lu, J.; Wang, H.; Qin, S.; Cao, L.; Pu, R.; Li, G.; Sun, J. Estimation of aboveground biomass of Robinia pseudoacacia forest in the Yellow River Delta based on UAV and Backpack LiDAR point clouds. Int. J. Appl. Earth Obs. Geoinf. 2020, 86, 102014. [Google Scholar] [CrossRef]

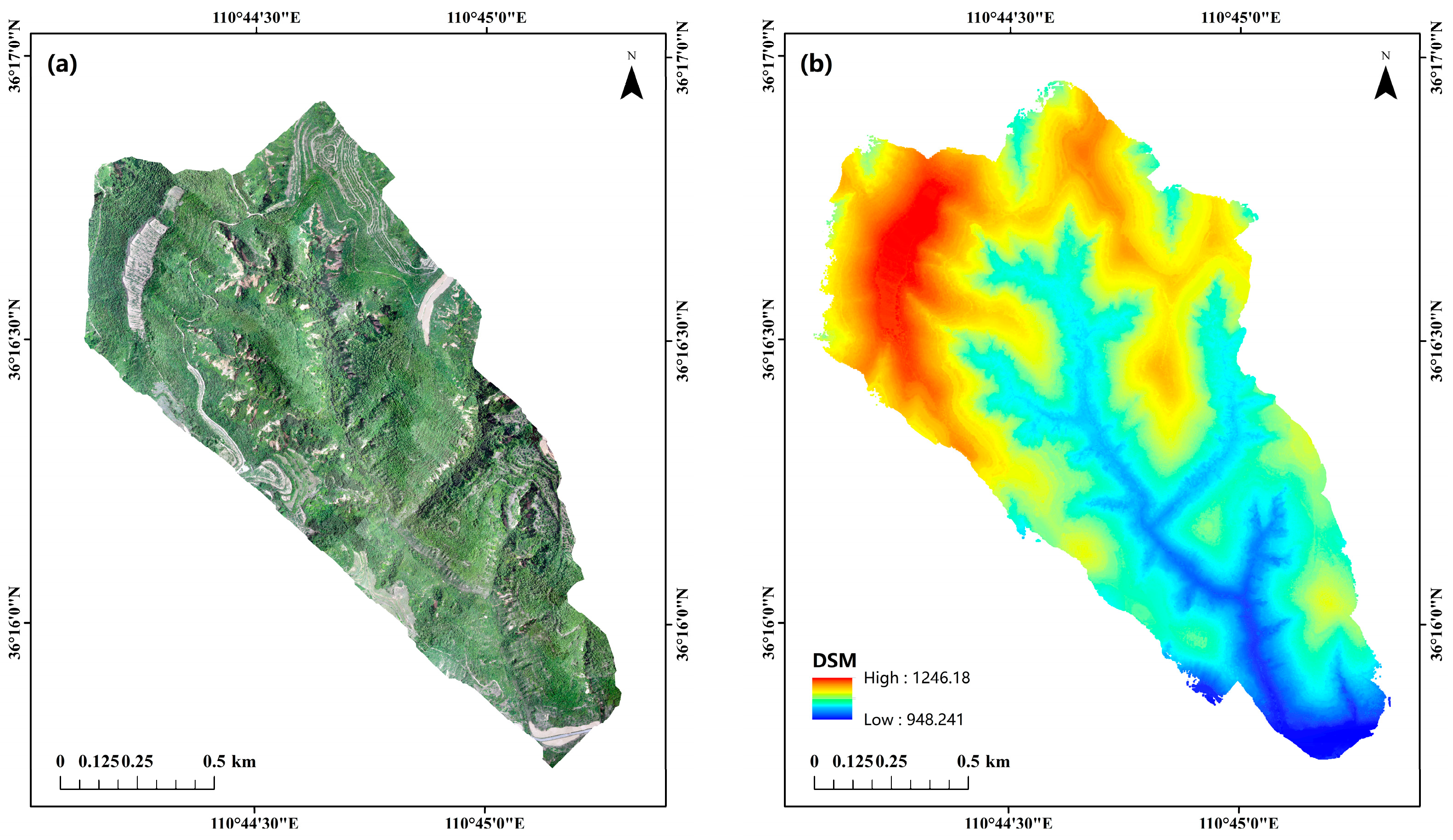
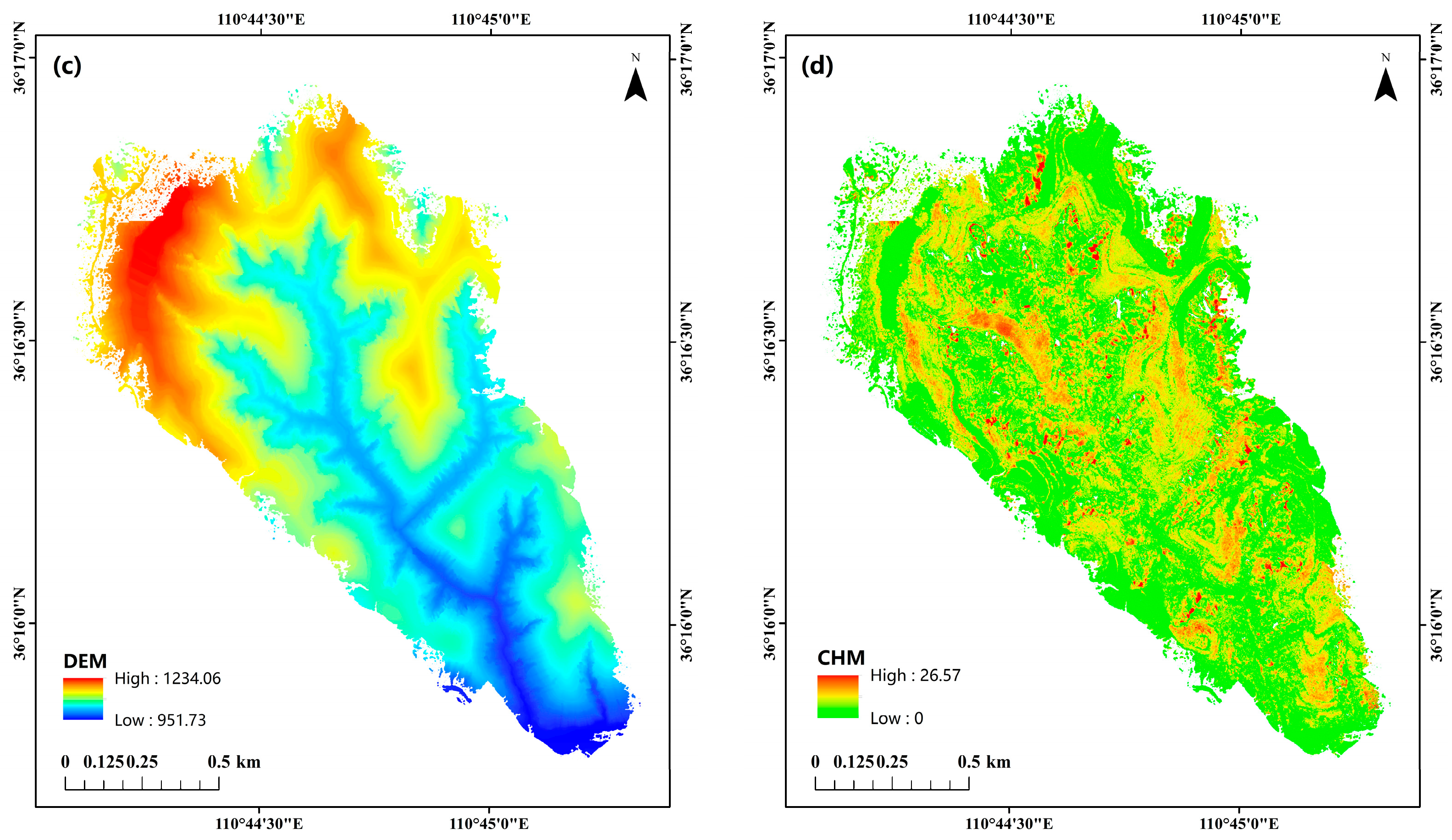


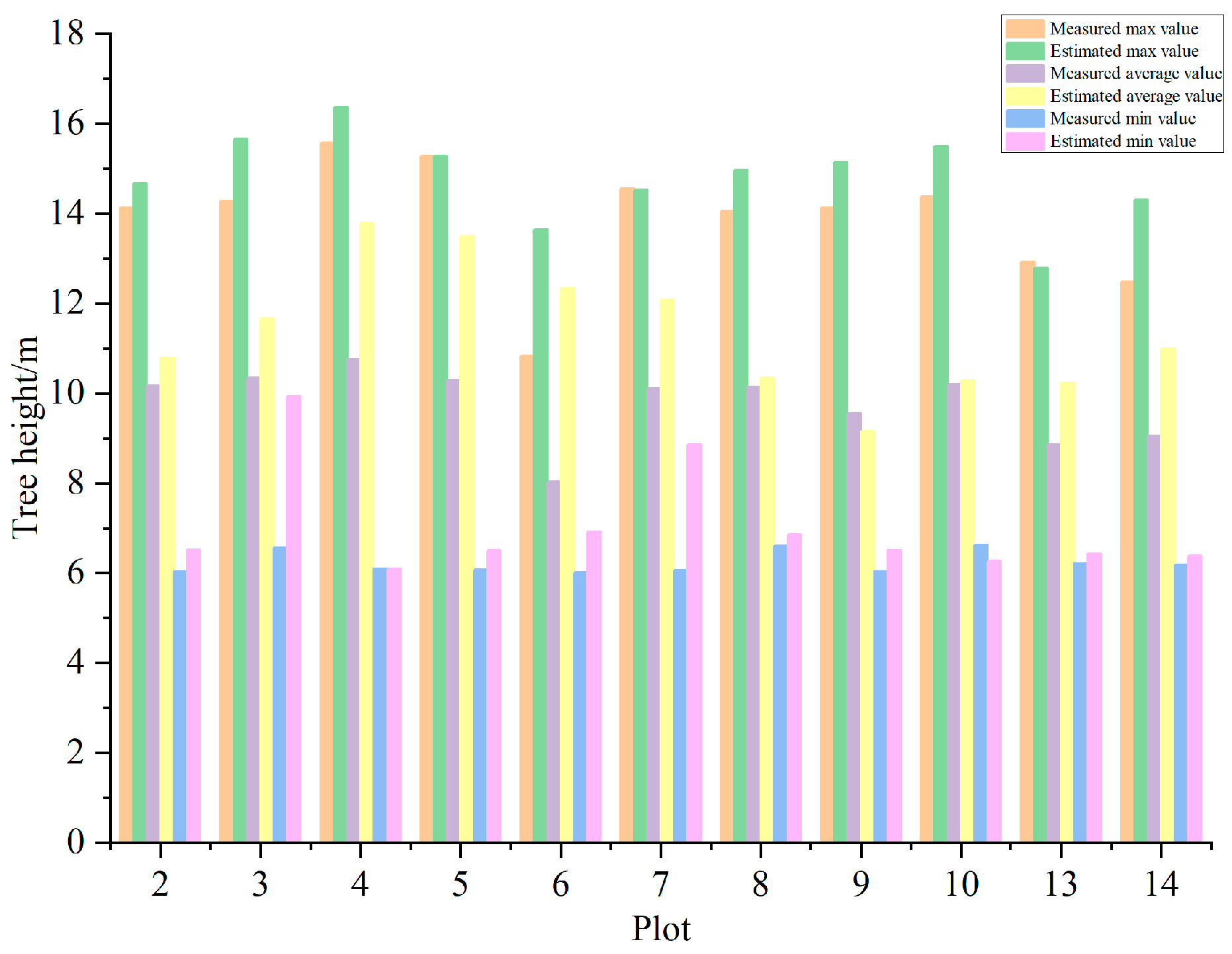


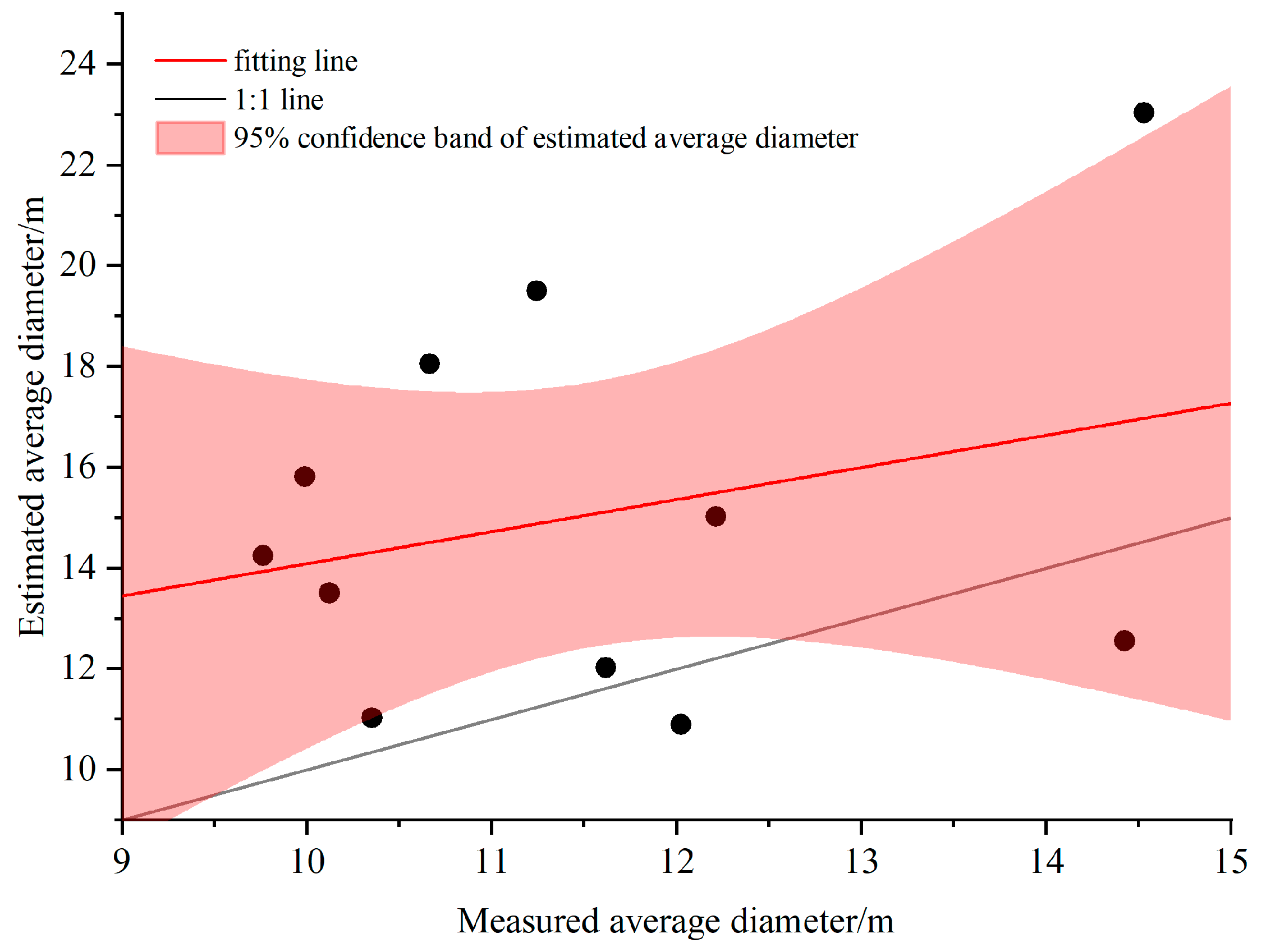
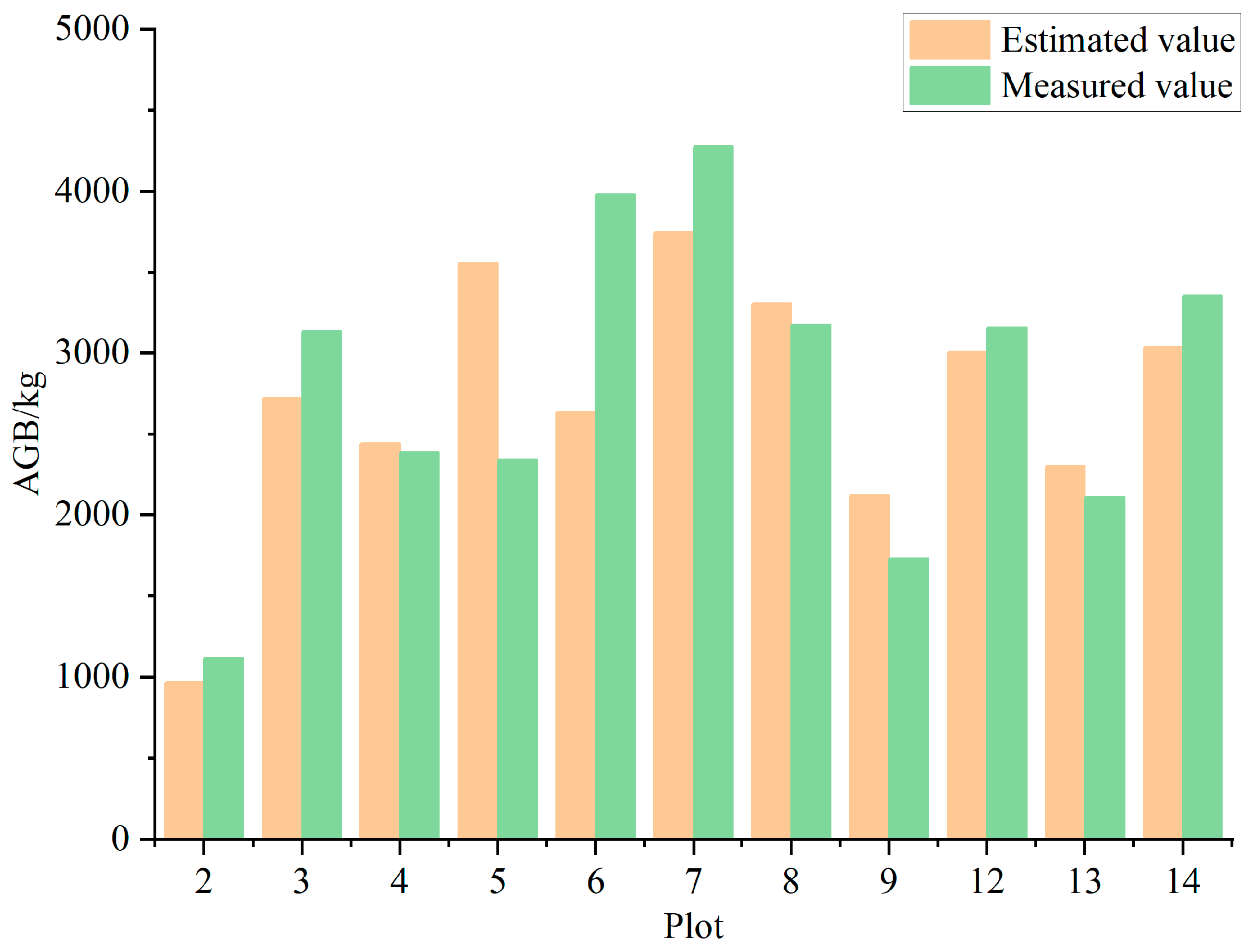
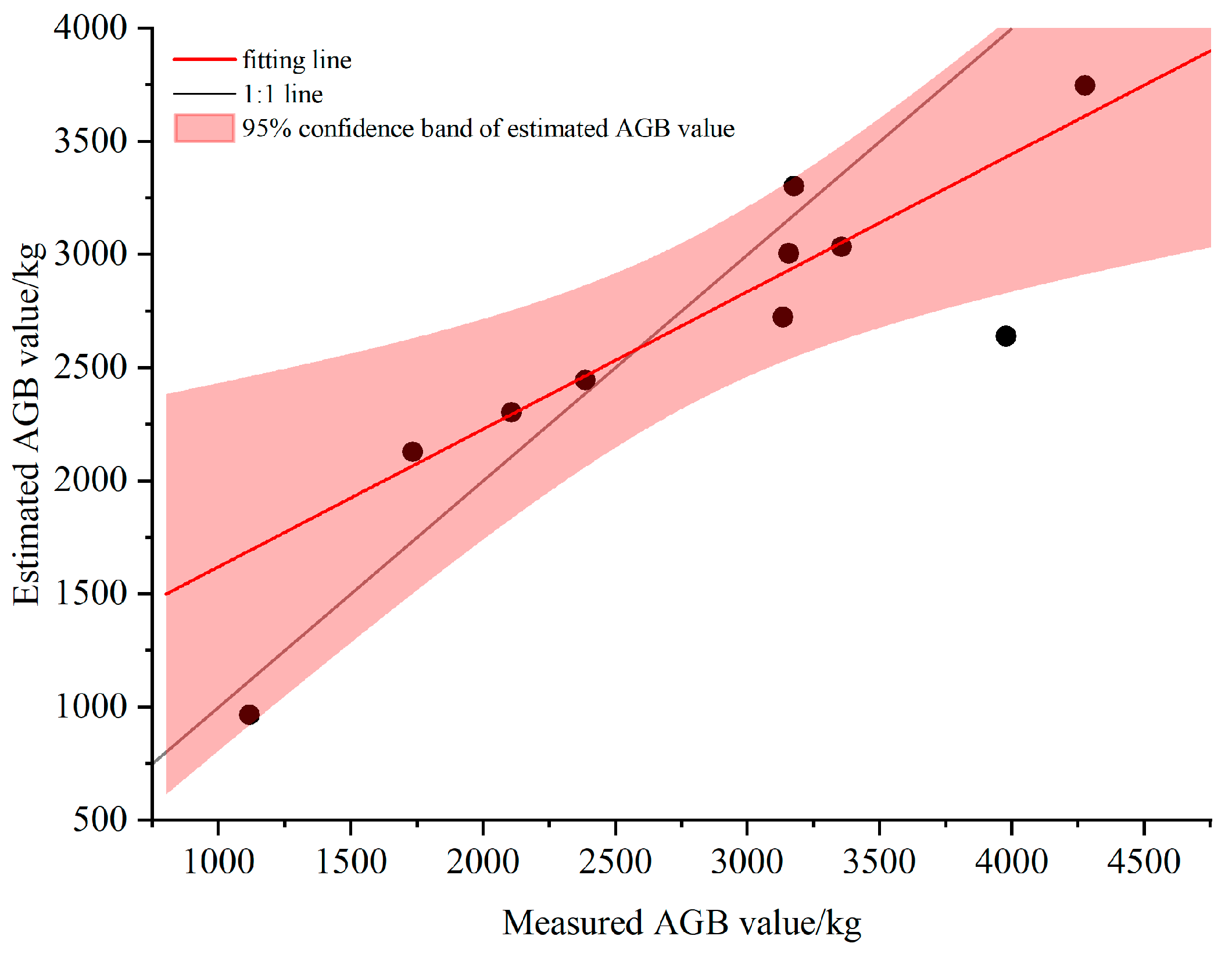
| Plot ID | Longitude | Latitude | Density (Tree/hm2) | Altitude/m | Aspect/° | Slope/° | Tree Height/m (avg. ± SD) | CV | AGB/kg |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 110°44′49.4810″ E | 36°16′28.2848″ N | 950 | 1181 | 335 | 25 | 11.46 ± 2.63 | Middle | 2107.996 |
| 2 | 110°45′27.1682″ E | 36°16′15.3416″ N | 1156 | 1112 | 65 | 27 | 10.17 ± 3.59 | High | 1117.346 |
| 3 | 110°44′24.8988″ E | 36°16′32.1703″ N | 1150 | 1166 | 180 | 20 | 10.34 ± 3.39 | High | 3134.514 |
| 4 | 110°45′26.5978″ E | 36°16′18.8931″ N | 1475 | 1119 | 75 | 26 | 10.77 ± 3.53 | High | 2386.459 |
| 5 | 110°45′50.7713″ E | 36°16′30.3138″ N | 1500 | 1117 | 60 | 15 | 8.27 ± 3.91 | High | 2344.06 |
| 6 | 110°44′31.3110″ E | 36°16′30.0838″ N | 1525 | 1149 | 225 | 23 | 8.03 ± 2.31 | Middle | 3978.908 |
| 7 | 110°46′11.1767″ E | 36°16′18.0881″ N | 1950 | 1022 | 70 | 31 | 10.1 ± 2.61 | Middle | 4275.538 |
| 8 | 110°46′08.4736″ E | 36°16′18.0780″ N | 2200 | 958 | 50 | 15 | 10.15 ± 2.34 | Middle | 3177.047 |
| 9 | 110°45′53.9493″ E | 36°16′29.8737″ N | 2050 | 1086 | 40 | 15 | 9.56 ± 2.54 | Middle | 1734.486 |
| 10 | 110°45′52.9134″ E | 36°16′32.7558″ N | 2444 | 1088 | 35 | 0 | 10.2 ± 2.19 | Middle | 2331.556 |
| 11 | 110°44′32.5029″ E | 36°16′42.0954″ N | 2475 | 1168 | 196 | 22 | 12.98 ± 3.03 | Middle | 3383.656 |
| 12 | 110°45′19.9705″ E | 36°16′52.4691″ N | 2475 | 1110 | 195 | 28 | 12.05 ± 3.63 | Middle | 3156.846 |
| 13 | 110°45′41.4187″ E | 36°16′20.3522″ N | 3022 | 1056 | 150 | 14 | 8.85 ± 2.85 | High | 2108.085 |
| 14 | 110°45′44.2842″ E | 36°16′22.4456″ N | 3300 | 1081 | 180 | 36 | 9.05 ± 2.68 | Middle | 3355.007 |
| Estimation Models | Fitting Formula | R2 | MAE | RMSE |
|---|---|---|---|---|
| AGB = aDBHb | AGB = 3.362 × DBH1.11 | 0.67 | 12.086 | 8.398 |
| AGB = aHb | AGB = 3.438 × H1.07 | 0.41 | 12.086 | 11.34 |
| AGB = a(DBH2 H)b | AGB = 2.111 × (DBH2 H)0.4333 | 0.70 | 4.514 | 8.188 |
| AGB = aDBHb Hc | AGB = 2.354 × DBH0.9392 H0.3171 | 0.69 | 11.595 | 8.595 |
| AGB = a(DBH3/H) b | AGB = 6.24 × (DBH3/H)0.4261 | 0.58 | 13.454 | 9.481 |
| Plot ID | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 13 | 14 | SUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reality | 60 | 31 | 44 | 32 | 63 | 52 | 22 | 49 | 34 | 64 | 63 | 514 |
| 0.1 × 0.1 m | 47 | 33 | 28 | 28 | 50 | 52 | 22 | 44 | 42 | 58 | 70 | 476 |
| Tp | 39 | 25 | 14 | 22 | 46 | 40 | 20 | 37 | 32 | 52 | 47 | 374 |
| Fn | 21 | 6 | 30 | 10 | 17 | 12 | 2 | 12 | 2 | 12 | 16 | 132 |
| Fp | 8 | 8 | 14 | 6 | 4 | 12 | 2 | 7 | 10 | 6 | 23 | 102 |
| r/% | 75.0 | 80.6 | 31.8 | 68.8 | 73.0 | 76.9 | 90.9 | 75.5 | 94.1 | 81.3 | 74.6 | 72.7 |
| p/% | 83.0 | 75.8 | 50.0 | 78.6 | 92.0 | 75.5 | 90.9 | 82.2 | 76.2 | 89.7 | 67.1 | 78.6 |
| 0.2 × 0.2 m | 40 | 32 | 25 | 30 | 45 | 46 | 21 | 40 | 38 | 54 | 64 | 435 |
| Tp | 32 | 24 | 11 | 24 | 41 | 33 | 19 | 32 | 28 | 48 | 41 | 333 |
| Fn | 28 | 7 | 33 | 8 | 22 | 19 | 3 | 17 | 6 | 16 | 22 | 173 |
| Fp | 8 | 8 | 14 | 6 | 4 | 13 | 2 | 8 | 10 | 6 | 23 | 102 |
| r/% | 61.5 | 77.4 | 25.0 | 75.0 | 65.1 | 63.5 | 86.4 | 65.3 | 82.4 | 75.0 | 65.1 | 64.7 |
| p/% | 80.0 | 75.0 | 44.0 | 80.0 | 91.1 | 71.7 | 90.5 | 80.0 | 73.7 | 88.9 | 64.1 | 76.6 |
| 0.3 × 0.3 m | 35 | 27 | 19 | 25 | 37 | 42 | 19 | 36 | 31 | 45 | 43 | 359 |
| Tp | 27 | 19 | 5 | 19 | 33 | 29 | 17 | 28 | 21 | 39 | 20 | 257 |
| Fn | 33 | 12 | 39 | 13 | 30 | 23 | 5 | 21 | 13 | 25 | 43 | 249 |
| Fp | 8 | 8 | 14 | 6 | 4 | 13 | 2 | 8 | 10 | 6 | 23 | 102 |
| r/% | 51.9 | 61.3 | 11.4 | 59.4 | 52.4 | 55.8 | 77.3 | 57.1 | 61.8 | 60.9 | 31.7 | 50.0 |
| p/% | 77.1 | 70.4 | 26.3 | 76.0 | 89.2 | 69.0 | 89.5 | 77.8 | 67.7 | 86.7 | 46.5 | 71.6 |
| 0.4 × 0.4 m | 32 | 23 | 18 | 22 | 29 | 36 | 15 | 31 | 22 | 38 | 30 | 296 |
| Tp | 24 | 15 | 4 | 16 | 29 | 26 | 13 | 23 | 21 | 33 | 24 | 228 |
| Fn | 36 | 16 | 40 | 16 | 34 | 26 | 9 | 26 | 13 | 31 | 39 | 278 |
| Fp | 8 | 8 | 14 | 6 | 0 | 10 | 2 | 8 | 1 | 5 | 6 | 68 |
| r/% | 46.2 | 48.4 | 9.1 | 50.0 | 46.0 | 50.0 | 59.1 | 46.9 | 61.8 | 51.6 | 38.1 | 44.3 |
| p/% | 75.0 | 65.2 | 22.2 | 72.7 | 100.0 | 72.2 | 86.7 | 74.2 | 95.5 | 86.8 | 80.0 | 77.0 |
| 0.5 × 0.5 m | 25 | 15 | 17 | 20 | 25 | 30 | 12 | 28 | 20 | 31 | 24 | 247 |
| Tp | 18 | 10 | 4 | 14 | 23 | 20 | 10 | 21 | 18 | 25 | 19 | 182 |
| Fn | 42 | 21 | 40 | 18 | 40 | 32 | 12 | 28 | 16 | 39 | 44 | 324 |
| Fp | 7 | 5 | 13 | 6 | 2 | 10 | 2 | 7 | 2 | 6 | 5 | 65 |
| r/% | 34.6 | 32.3 | 9.1 | 43.8 | 36.5 | 38.5 | 42.5 | 42.9 | 52.9 | 39.1 | 30.2 | 35.4 |
| p/% | 72.0 | 66.7 | 23.5 | 70.0 | 92.0 | 66.7 | 83.3 | 75.0 | 90.0 | 80.6 | 79.2 | 73.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Zhang, X.; Zhang, J.; Zhang, Y.; Hu, Y.; Zhao, J.; Li, Y. Estimating the Aboveground Biomass of Robinia pseudoacacia Based on UAV LiDAR Data. Forests 2024, 15, 548. https://doi.org/10.3390/f15030548
Cheng J, Zhang X, Zhang J, Zhang Y, Hu Y, Zhao J, Li Y. Estimating the Aboveground Biomass of Robinia pseudoacacia Based on UAV LiDAR Data. Forests. 2024; 15(3):548. https://doi.org/10.3390/f15030548
Chicago/Turabian StyleCheng, Jiaqi, Xuexia Zhang, Jianjun Zhang, Yanni Zhang, Yawei Hu, Jiongchang Zhao, and Yang Li. 2024. "Estimating the Aboveground Biomass of Robinia pseudoacacia Based on UAV LiDAR Data" Forests 15, no. 3: 548. https://doi.org/10.3390/f15030548
APA StyleCheng, J., Zhang, X., Zhang, J., Zhang, Y., Hu, Y., Zhao, J., & Li, Y. (2024). Estimating the Aboveground Biomass of Robinia pseudoacacia Based on UAV LiDAR Data. Forests, 15(3), 548. https://doi.org/10.3390/f15030548






