The Utilization and Roles of Nitrogen in Plants
Abstract
1. Introduction
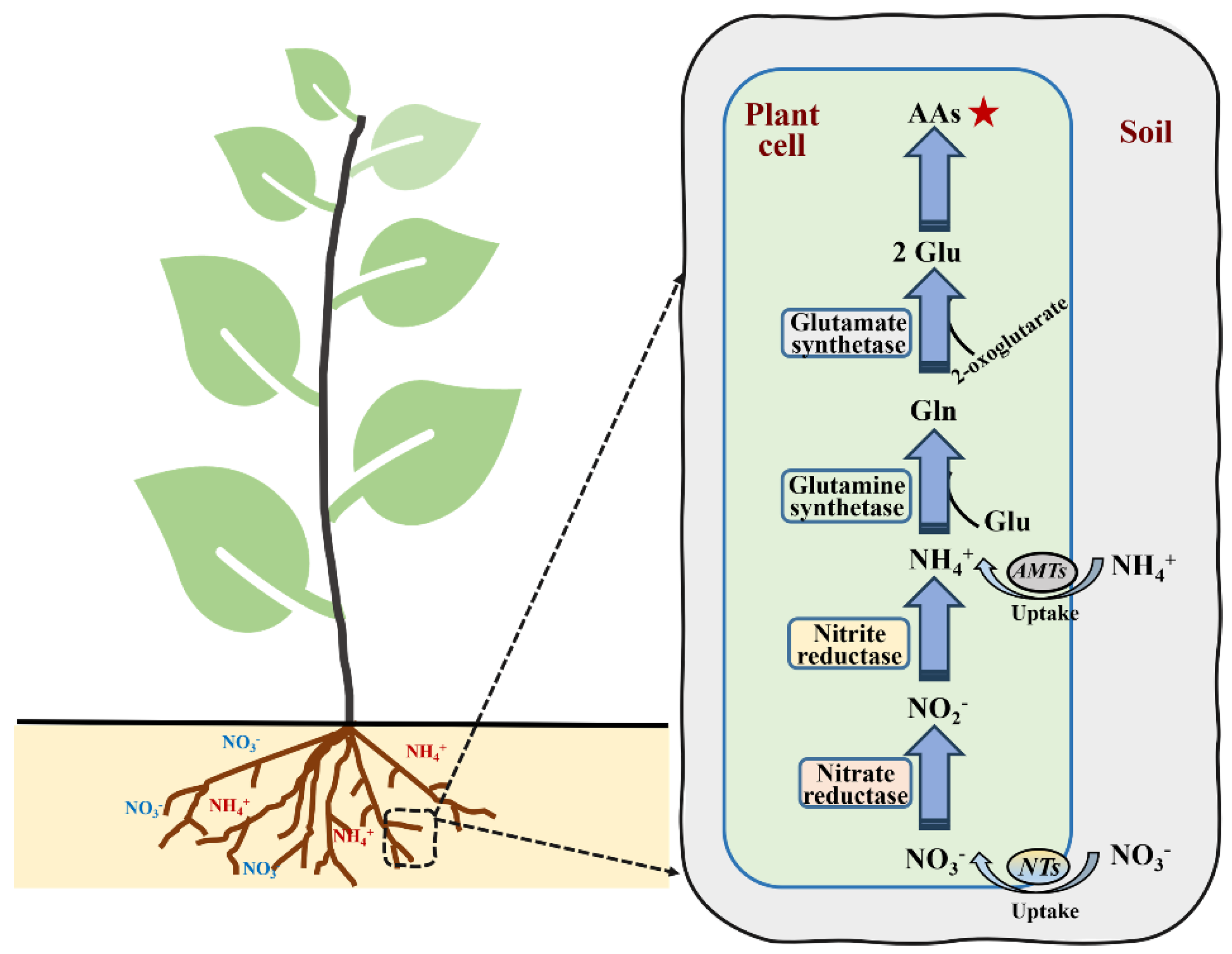
2. Study of the Mechanism of N Uptake and Assimilation in Plants
2.1. Research Progress on NO3− Uptake in Plants
2.2. Research Progress on NH4+ Uptake in Plants
2.3. Nitrogen Assimilation in Plants
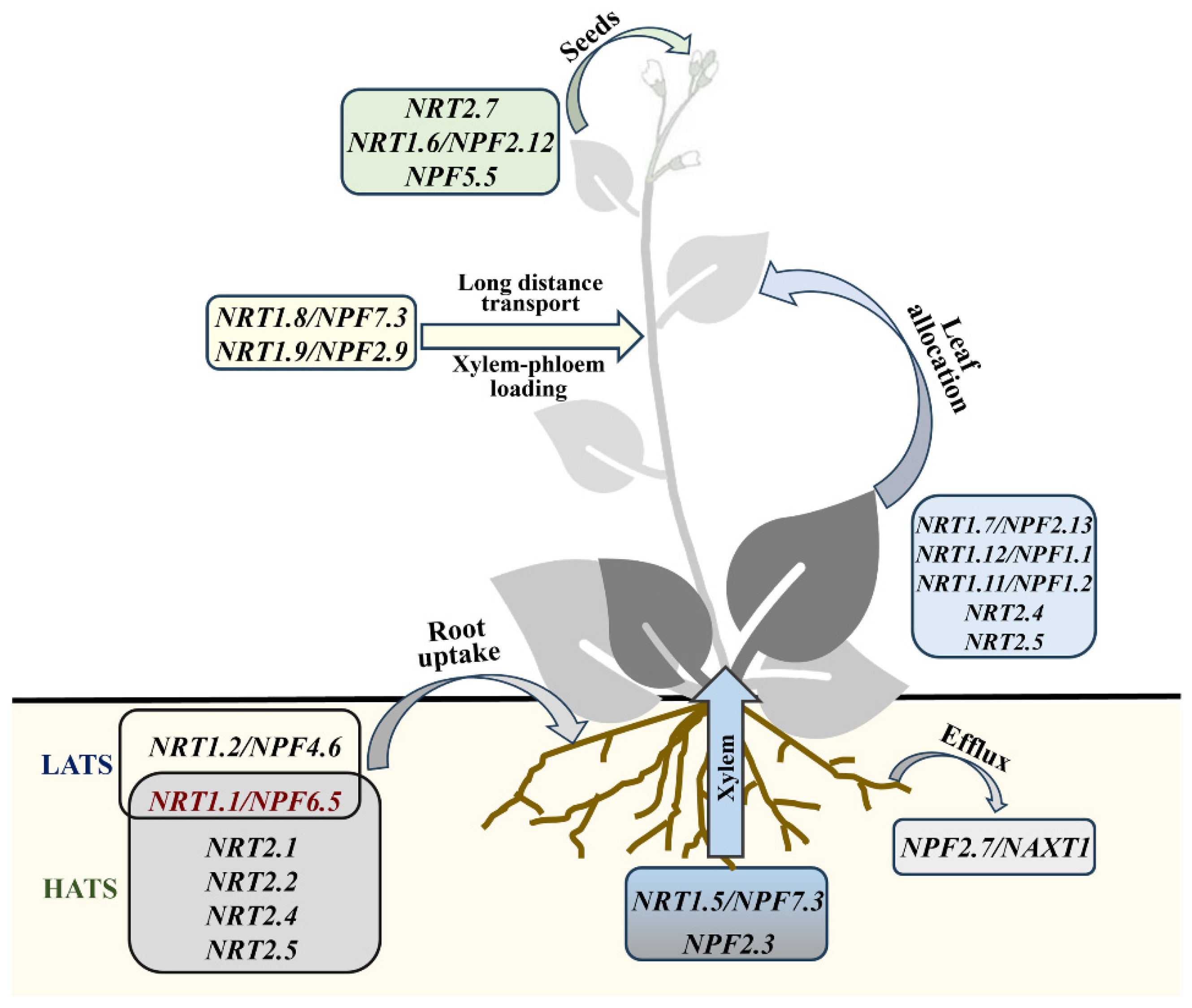
3. Research Progress on Plants’ N Utilization and Regulation
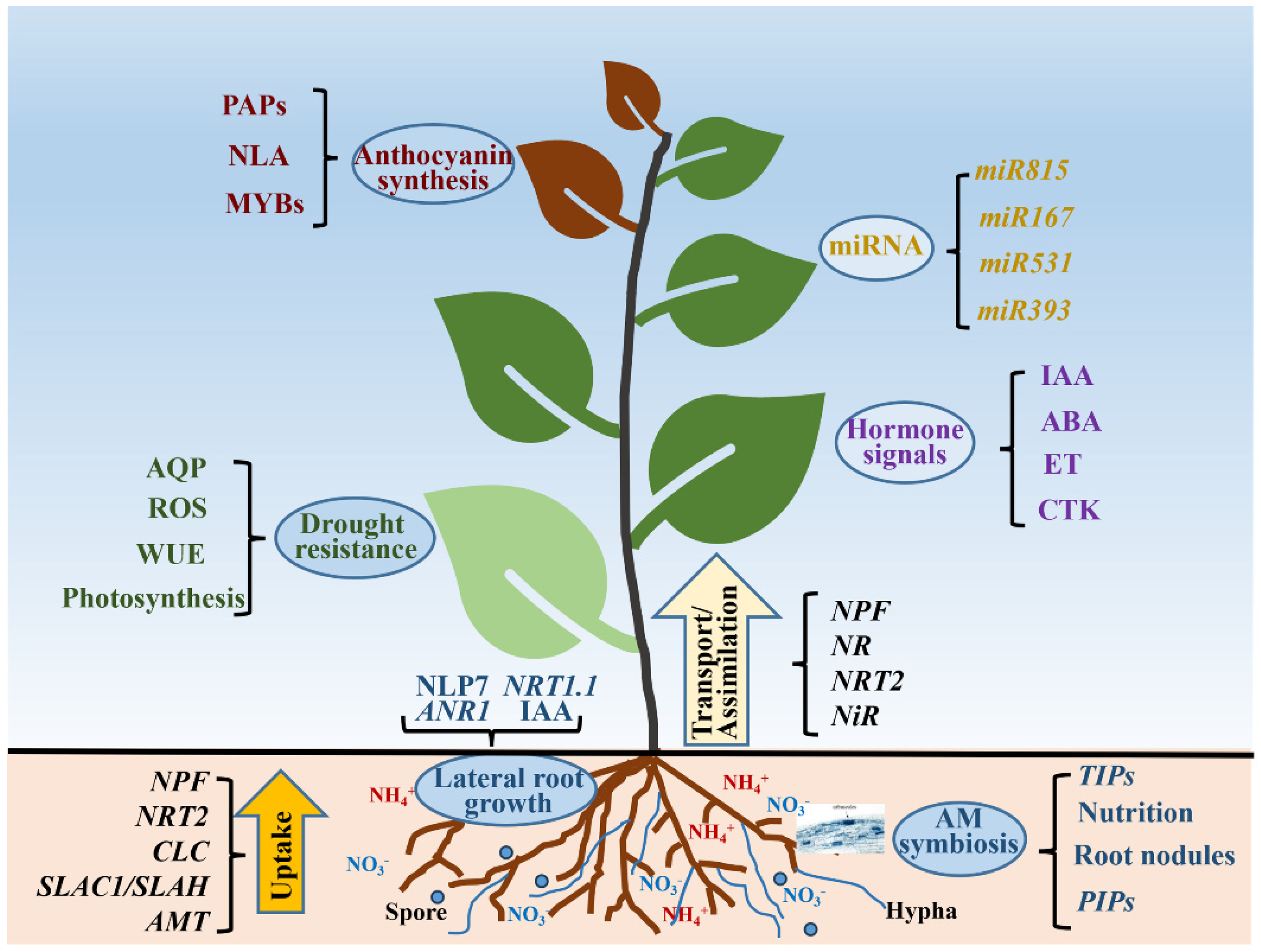
4. Regulation Roles of Nitrogen in Plants
4.1. Nitrogen and Hormone Signals
4.2. Nitrogen Metabolism and miRNA Regulation
4.3. Nitrogen Regulates Lateral Root Growth
4.4. Nitrogen and Drought Resistance
4.5. Nitrogen Regulates Anthocyanin Synthesis
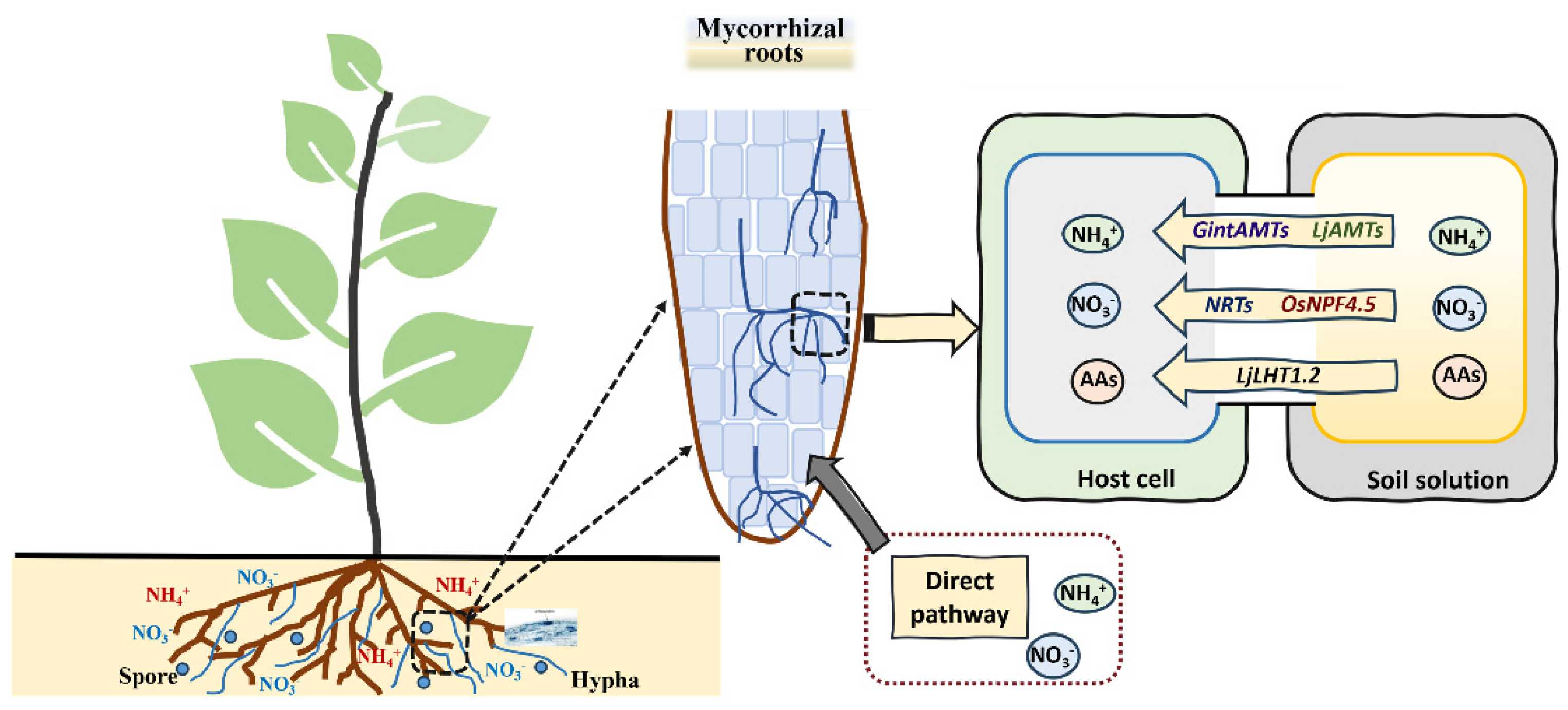
4.6. Nitrogen and Mycorrhizal Symbiosis
4.7. Nitrogen and Rhizobium–Legume Symbiosis
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- O′Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Q.; Wang, X.; Zhang, J.; Ismail, A.M.; Zhang, Z. Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5. Plant Cell Environ. 2021, 44, 3806–3818. [Google Scholar] [CrossRef]
- Jiang, H.; Qi, C.H.; Gao, H.N.; Feng, Z.Q.; Wu, Y.T.; Xu, X.X.; Cui, J.Y.; Wang, X.F.; Lv, Y.H.; Gao, W.S.; et al. MdBT2 regulates nitrogen-mediated cuticular wax biosynthesis via a MdMYB106-MdCER2L1 signalling pathway in apple. Nat. Plants 2024, 10, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ma, Z.; Li, J.; Bian, N.; Guo, Z.; Guo, J.; Zhao, S.; Chu, B.; Niu, C.; Ma, F.; et al. Interfering small ubiquitin modifiers (SUMO) exhibits apple’s enhanced tolerance to nitrogen deficiency. Fruit Res. 2023, 3, 24. [Google Scholar] [CrossRef]
- Nacry, P.; Bouguyon, E.; Gojon, A. Nitrogen acquisition by roots: Physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 2013, 370, 1–29. [Google Scholar] [CrossRef]
- Luo, L.; Zhao, C.; Zheng, D.; Wang, E.; Liang, J.; Yin, C. Nitrogen uptake preference and allocation in Populus cathayana in response to drought stress. Environ. Exp. Bot. 2023, 213, 105415. [Google Scholar] [CrossRef]
- Ma, Q.; Zhao, C.; Hu, S.; Zuo, K. Arabidopsis calcium-dependent protein kinase CPK6 regulates drought tolerance under high nitrogen by the phosphorylation of NRT1.1. J. Exp. Bot. 2023, 74, 5682–5693. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ma, W.; Song, J.; Lu, M.; Rahman, S.; Bui, T.T.X.; Vu, D.D.; Zheng, H.; Wang, J.; Zhang, Y. Physiological and transcriptional responses of Catalpa bungei to drought stress under sufficient- and deficient-nitrogen conditions. Tree Physiol. 2017, 247, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Cao, X.; Zhang, M. Plant nitrogen availability and crosstalk with phytohormones signallings and their biotechnology breeding application in crops. Plant Biotechnol. J. 2023, 21, 1320–1342. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A. Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr. Opin. Plant Biol. 2015, 25, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Liu, M.; Lin, Z.; Wang, Z.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Chaput, V.; Przybyla-Toscano, J.; Fayos, I.; Ibarra, C.; Moyano, T.; Fizames, C.; Tillard, P.; O’Brien, J.A.; Gutiérrez, R.A.; et al. Genome-wide analysis in response to nitrogen and carbon identifies regulators for root AtNRT2 transporters. Plant Physiol. 2021, 186, 696–714. [Google Scholar] [CrossRef] [PubMed]
- Léran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of Nitrate Transporter 1/Peptide Transporter family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, B.; Li, A.; Chu, C. NRT1.1s in plants: Functions beyond nitrate transport. J. Exp. Bot. 2020, 71, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, T.J. CLC chloride channels and transporters, from genes to protein structure, pathology and physiology. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 3–36. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, X.; Ye, S.; Zheng, J.; Huang, X.; Yu, F.; Chen, Z.; Cai, S.; Zhang, P. Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids. Nat. Commun. 2023, 14, 4879. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Song, H.; Liao, Q.; Yu, Y.; Jian, S.; Lepo, J.E.; Liu, Q.; Rong, X.; Tian, C.; Zeng, J.; et al. Nitrogen use efficiency is mediated by vacuolar nitrate sequestration capacity in roots of Brassica napus. Plant Physiol. 2016, 170, 1684–1698. [Google Scholar] [CrossRef] [PubMed]
- Hodin, J.; Lind, C.; Marmagne, A.; Espagne, C.; Bianchi, M.W.; Angeli, A.D.; Abou-Choucha, F.; Bourge, M.; Chardon, F.; Thomine, S.; et al. Proton exchange by the vacuolar nitrate transporter CLCa is required for plant growth and nitrogen use efficiency. Plant Cell 2022, 35, 318–335. [Google Scholar] [CrossRef] [PubMed]
- Fecht-Bartenbach, J.; Bogner, M.; Dynowski, M.; Ludewig, U. CLCb-mediated NO3−/H+ exchange across the tonoplast of Arabidopsis vacuoles. Plant Cell Physiol. 2010, 51, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, D.; He, Y.; Tang, J.; Chen, H.; Gong, P.; Luo, J.; Zhang, Z. CHLORIDE CHANNEL-b mediates vacuolar nitrate efflux to improve low nitrogen adaptation in Arabidopsis. Plant Physiol. 2023, 193, 1987–2002. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Fang, X.; Xiao, C.; Ma, Z.; Huang, X.; Su, J.; Li, J.; Wang, J.; Wang, S.; Luan, S.; et al. Kinase SnRK1.1 regulates nitrate channel SLAH3 engaged in nitrate-dependent alleviation of ammonium toxicity. Plant Physiol. 2021, 186, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Maierhofer, T.; Lind, C.; Huttl, S.; Scherzer, S.; Papenfuss, M.; Simon, J.; Al-Rasheid, K.A.; Ache, P.; Rennenberg, H.; Hedrich, R.; et al. A single-pore residue renders the Arabidopsis root anion channel SLAH2 highly nitrate selective. Plant Cell 2014, 26, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Geiger, D.; Maierhofer, T.; Al-Rasheid, K.A.; Scherzer, S.; Mumm, P.; Liese, A.; Ache, P.; Wellmann, C.; Marten, I.; Grill, E. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 2011, 4, ra32. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Sun, D.; Liu, B.; Fang, X.; Li, P.; Jiang, Y.; He, M.; Li, J.; Luan, S.; He, K. Nitrate transporter NRT1.1 and anion channel SLAH3 form a functional unit to regulate nitrate-dependent alleviation of ammonium toxicity. J. Integr. Plant Biol. 2022, 64, 942–957. [Google Scholar] [CrossRef]
- Jung, J.H.; Li, Z.; Chen, H.; Yang, S.; Li, D.; Priatama, R.A.; Kumar, V.; Xuan, Y.H. Mutation of phytochrome B promotes resistance to sheath blight and saline-alkaline stress via increasing ammonium uptake in rice. Plant J. 2023, 113, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Ninnemann, O.; Jauniaux, J.C.; Frommer, W.B. Identification of a high affinity NH4+ transporter from plants. EMBO J. 1994, 13, 3464–3471. [Google Scholar] [CrossRef]
- Giehl, R.; Laginha, A.; Duan, F.; Rentsch, D.; Yuan, L.; Wirén, N. A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Mol. Plant 2017, 10, 1449–1460. [Google Scholar] [CrossRef]
- Suenaga, A.; Moriya, K.; Sonoda, Y.; Ikeda, A.; Wiren, N.; Hayakawa, T.; Yamaguchi, J.; Yamaya, T. Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 2003, 44, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Tami, M.; Fred, D.; Franois, L. Multiple horizontal gene transfers of ammonium transporters/ammonia permeases from prokaryotes to eukaryotes: Toward a new functional and evolutionary classification. Mol. Biol. Evol. 2011, 29, 51–60. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, W.; Wu, J.; Xie, K.; Li, X. LjAMT2;2 promotes ammonium nitrogen transport during arbuscular mycorrhizal fungi symbiosis in Lotus japonicus. Int. J. Mol. Sci. 2022, 23, 9522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhao, Y.; Luo, W.; Li, R.; He, Q.; Fang, X.; Michele, R.D.; Ast, C.; Wirén, N.V.; Lin, J. Single-particle analysis reveals shutoff control of the Arabidopsis ammonium transporter AMT1;3 by clustering and internalization. Proc. Natl. Acad. Sci. USA 2013, 110, 13204–13209. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, Y.; Lai, L.; Liu, X.; Miao, C.; Liu, R.; Li, X.; Tan, J.; Gao, Z.; Chen, J. OsAMT1.1 expression by nitrate-inducible promoter of OsNAR2.1 increases nitrogen use efficiency and rice yield. Rice Sci. 2023, 30, 222–234. [Google Scholar] [CrossRef]
- Yuan, L.; Graff, L.; Loqué, D.; Kojima, S.; Tsuchiya, Y.N.; Takahashi, H.; Wirén, N. AtAMT1;4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis. Plant Cell Physiol. 2009, 50, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Transporters involved in mineral nutrient uptake in rice. J. Exp. Bot. 2016, 67, 3645–3653. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Yue, X.; Fang, S.; Qian, M.; Zhou, S.; Shang, X.; Yang, W. Responses of nitrogen metabolism, photosynthetic parameter and growth to nitrogen fertilization in Cyclocarya paliurus. For. Ecol. Manag. 2021, 502, 119715. [Google Scholar] [CrossRef]
- Huang, W.T.; Zheng, Z.C.; Hua, D.; Chen, X.F.; Zhang, J.; Chen, H.H.; Ye, X.; Guo, J.X.; Yang, L.T.; Chen, L.S. Adaptive responses of carbon and nitrogen metabolisms to nitrogen-deficiency in Citrus sinensis seedlings. BMC Plant Biol. 2022, 22, 370. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, K.; Yamaya, T.; Hayakawa, T.; Mae, T.; Ojima, K. Vascular bundle-specific localization of cytosolic glutamine synthetase in rice leaves. Plant Physiol. 1992, 99, 1481–1486. [Google Scholar] [CrossRef]
- Avila-Ospina, L.; Marmagne, A.; Talbotec, J.; Krupinska, K.; Masclaux-Daubresse, C. The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. J. Exp. Bot. 2015, 66, 2013–2026. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Reisdorf-Cren, M.; Pageau, K.; Lelandais, M.; Grandjean, O.; Kronenberger, J.; Valadier, M.H.; Feraud, M.; Jouglet, T.; Suzuki, A. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 2006, 140, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, C.; Huang, D.; Dong, A.; Li, P.; Ma, F. High-efficient utilization and uptake of N contribute to higher NUE of ‘Qinguan’ apple under drought and N-deficient conditions compared with ‘Honeycrisp’. Tree Physiol. 2019, 39, 1880–1895. [Google Scholar] [CrossRef]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Chung, H.S.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Durand, M.; Brehaut, V.; Clement, G.; Kelemen, Z.; Macé, J.; Feil, R.; Duville, G.; Launay-Avon, A.; Roux, C.P.L.; Lunn, J.E.; et al. The Arabidopsis transcription factor NLP2 regulates early nitrate responses and integrates nitrate assimilation with energy and carbon skeleton supply. Plant Cell 2023, 35, 1429–1454. [Google Scholar] [CrossRef] [PubMed]
- Bouguyon, E.; Perrine-Walker, F.; Pervent, M.; Bach, L.; Krouk, G.; Gojon, A.; Nacry, P. Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiol. 2016, 172, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Tsay, Y.F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003, 22, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Bellegarde, F.; Gojon, A.; Martin, A. Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2553–2565. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [PubMed]
- Medici, A.; Marshall-Colon, A.; Ronzier, E.; Szponarski, W.; Wang, R.C.; Gojon, A.; Crawford, N.M.; Ruffe, S.; Coruzzi, G.M.; Krouk, G. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Li, T.; Wang, X.; Sun, W.; Zhang, T.; You, C.; Wang, X. Identification and characterization of apple MdNLP7 transcription factor in the nitrate response. Plant Sci. 2022, 316, 111158. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Liu, Z.; Guo, J.; Jia, Z.; Shi, Y.; Kang, K.; Peng, W.; Wang, Z.; Chen, L.; Neuhaeuser, B.; et al. ZmNRT1.1B (ZmNPF6.6) determines nitrogen use efficiency via regulation of nitrate transport and signalling in maize. Plant Biotechnol. J. 2024, 22, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Armijo, G.; Gutiérrez, R.A. Emerging players in the nitrate signaling pathway. Mol. Plant 2017, 10, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Ródenas, R.; Vert, G. Regulation of root nutrient transporters by CIPK23: ‘One kinase to rule them all’. Plant Cell Physiol. 2021, 62, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, M.; Yang, J.; Li, M.; Zhang, Z.; Gao, H.; Wang, C.; Tian, H. Brassinosteroid transcription factor BES1 modulates nitrate deficiency by promoting NRT2.1 and NRT2.2 transcription in Arabidopsis. Plant J. 2023, 114, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, Z.; Sun, H.; Liu, X.; Xie, J.; Qiu, Y.; Chai, T.; Chu, C.; Hu, B. Nitrate confers rice adaptation to high ammonium by suppressing its uptake but promoting its assimilation. Mol. Plant 2023, 16, 1871–1874. [Google Scholar] [CrossRef] [PubMed]
- Léran, S.; Edel, K.H.; Pervent, M.; Hashimoto, K.; Corratge-Faillie, C.; Offenborn, J.N.; Tillard, P.; Gojon, A.; Kudla, J.; Lacombe, B. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 2015, 8, ra43. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Duan, Y.; Ye, Q.; Ma, Y.; Ma, R.; Zhao, L.; Zhu, S.; Yu, F.; Qi, S.; Wang, Y. The Arabidopsis eIF4E1 regulates NRT1.1-mediated nitrate signaling at both translational and transcriptional levels. New Phytol. 2023, 240, 338–353. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, Q.; Ye, Y.; Li, T.; Sun, X.; Huo, L.; Wang, P.; Gong, X.; Ma, F. MdATG5a positively regulates nitrogen uptake under low nitrogen conditions by enhancing the accumulation of flavonoids and auxin in apple roots. Environ. Exp. Bot. 2022, 197, 104840. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Trivellini, A.; Fatma, M.; Masood, A.; Francini, A.; Iqbal, N.; Ferrante, A.; Khan, N.A. Role of ethylene in responses of plants to nitrogen availability. Front. Plant Sci. 2015, 6, 927. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Ma, T.; Xian, W.; Hu, B.; Chu, C. Interplay between ethylene and nitrogen nutrition: How ethylene orchestrates nitrogen responses in plants. J. Integr. Plant Biol. 2023, 65, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Guinel, F.C. Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 2015, 6, 1121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yi, H.; Gong, J. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 2014, 26, 3984–3998. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Sun, P.; Zhang, W. Ethylene is involved in nitrate-depend root growth and branching in Arabidopsis thaliana. New Phytol. 2009, 184, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Trivellini, A.; Masood, A.; Ferrante, A.; Khan, N.A. Current understanding on ethylene signaling in plants: The influence of nutrient availability. Plant Physiol. Biochem. 2013, 73, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Gernier, H.D.; Pessemier, J.D.; Xu, J.; Cristescu, S.M.; Straeten, D.V.D.; Verbruggen, N.; Hermans, C. A comparative study of ethylene emanation upon nitrogen deficiency in natural accessions of Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wang, Q.; Jing, G.; Ma, M.; Li, C.; Ma, F. Overexpression of MdIAA24 improves apple drought resistance by positively regulating strigolactone biosynthesis and mycorrhization. Tree Physiol. 2021, 41, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene TAR2 is involved in low nitrogen mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Qi, T.; Hu, Z.; Fan, X.; Zhu, L.; Iqbal, M.; Yin, X.; Xu, G.; Fan, X. OsNAR2.1 positively regulates drought tolerance and grain yield under drought stress conditions in rice. Front. Plant Sci. 2019, 10, 197. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Li, Z.; Waadt, R.; Schroeder, J.I. SnapShot: Abscisic acid signaling. Cell 2017, 171, 1708–1708.e1. [Google Scholar] [CrossRef]
- Ondzighi-Assoume, C.A.; Chakraborty, S.; Harris, J.M. Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. Plant Cell 2016, 28, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Wang, T.; Ju, C.; Deng, J.; Zhang, T.; Li, M.; Tian, H.; Wang, C. Abscisic acid signaling negatively regulates nitrate uptake via phosphorylation of NRT1.1 by SnRK2s in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, P.; Liu, Q.; Li, G.; Di, D.; Xia, G.; Kronzucker, H.J.; Fang, S.; Chu, J.; Shi, W. TaANR1-TaBG1 and TaWabi5-TaNRT2s/NARs link ABA metabolism and nitrate acquisition in wheat roots. Plant Physiol. 2020, 182, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 2021, 105, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xing, X.; Wang, Y.; Tran, A.; Crawford, N.M. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 2009, 151, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef] [PubMed]
- Hervé, C.; Dabos, P.; Bardet, C.; Jauneau, A.; Auriac, M.C.; Ramboer, A.; Lacout, F.; Tremousaygue, D. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009, 149, 1462–1477. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.; Du, C.; Gao, H.; Liu, S.; Sun, W.; Lu, H.; Kang, J.; Xie, Y.; Ma, D.; Wang, C. Identification of microRNAs in developing wheat grain that are potentially involved in regulating grain characteristics and the response to nitrogen levels. BMC Plant Biol. 2020, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Y.; Chai, X.; Foster, T.M.; Deng, C.H.; Wu, T.; Zhang, X.; Han, Z.; Wang, Y. miR164-MhNAC1 regulates apple root nitrogen uptake under low nitrogen stress. New Phytol. 2024, 242, 1218–1237. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, D.L.; Paola, D.D.; Janni, M.; Curci, P.L.; Sonnante, G. Durum wheat miRNAs in response to nitrogen starvation at the grain filling stage. PLoS ONE 2017, 12, e0183253. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Estelle, M. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, Y.; Cao, Y.; Chen, H.; Wang, J.; Bi, Y.; Tian, F.; Yang, F.; Rothstein, S.J.; Zhou, X.; et al. Overexpression of miR169o, an overlapping MicroRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice. Plant Cell Physiol. 2018, 59, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.J.; Beatty, P.H.; Good, A.G.; Muench, D.G. Manipulation of microRNA expression to improve nitrogen use efficiency. Plant Sci. 2013, 210, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; Macpherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, H.N. IAA-Ala resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Musialaklange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.R. Identification of nutrient-responsive Arabidopsis and rapeseed micro RNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wu, F.; Lin, Y.; Han, X.; Xu, X.; Zhang, Y.; Yang, Q.; Huang, H.; Tong, Z.; Zhang, J. A miR169c-NFYA10 module confers tolerance to low-nitrogen stress to Betula luminifera. Ind. Crops Prod. 2021, 172, 13988. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Sagwal, V.; Sihag, P.; Singh, Y.; Mehla, S.; Kapoor, P.; Balyan, P.; Kumar, A.; Mir, R.R.; Dhankher, O.P.; Kumar, U. Development and characterization of nitrogen and phosphorus use efficiency responsive genic and miRNA derived SSR markers in wheat. Heredity 2022, 128, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Bellegarde, F.; Sakakibara, H. Nitrate-dependent modulation of root system architecture in maize: A balance between strigolactone and auxin pathways. Plant Cell Physiol. 2021, 62, 541–542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gong, X.; Cheng, S.; Yu, H.; Li, D.; Su, X.; Lei, Z.; Li, M.; Ma, F. Proline-rich protein MdPRP6 alters low nitrogen stress tolerance by regulating lateral root formation and anthocyanin accumulation in transgenic apple (Malus domestica). Environ. Exp. Bot. 2022, 197, 104841. [Google Scholar] [CrossRef]
- Tahir, M.M.; He, X.; Liu, Y.; Raza, H.; Aziz, U.; Fan, L.; Asghar, Z.; Li, S.; Sun, S.; Zhang, D.; et al. Nitrate stimulates adventitious rooting by increasing auxin content and regulating auxin- and root development-related genes expression in apple. Hortic. Adv. 2023, 1, 18. [Google Scholar] [CrossRef]
- Rellan-Alvarez, R.; Lobet, G.; Dinneny, J.R. Environmental control of root system biology. Annu. Rev. Plant Biol. 2016, 67, 619–642. [Google Scholar] [CrossRef]
- Tokizawa, M.; Enomoto, T.; Chandnani, R.; Mora-Macías, J.; Burbridge, C.; Armenta-Medina, A.; Kobayashi, Y.; Yamamoto, Y.Y.; Koyama, H.; Kochian, L.V. The transcription factors, STOP1 and TCP20, are required for root system architecture alterations in response to nitrate deficiency. Proc. Natl. Acad. Sci. USA 2023, 120, e2300446120. [Google Scholar] [CrossRef] [PubMed]
- Contreras-López, O.; Vidal, E.A.; Riveras, E.; Alvarez, J.M.; Moyano, T.C.; Sparks, E.E.; Medina, J.; Pasquino, A.; Benfey, P.N.; Coruzzi, G.M.; et al. Spatiotemporal analysis identifies ABF2 and ABF3 as key hubs of endodermal response to nitrate. Proc. Natl. Acad. Sci. USA 2022, 119, e2107879119. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhu, M.; Jia, L.; Xie, Y.; Wang, Z.; Xuan, W. Ammonium transporters cooperatively regulate rice crown root formation responding to ammonium nitrogen. J. Exp. Bot. 2022, 73, 3671–3685. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Krall, L.; Li, Z.; Xi, L.; Luo, H.; Li, S.; He, M.; Yang, X.; Zan, H.; Gilbert, M.; et al. Transceptor NRT1.1 and receptor-kinase QSK1 complex controls PM H+-ATPase activity under low nitrate. Curr. Biol. 2024, 34, 1479–1491.e6. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ji, Z.; Tao, Y.; Wei, S.; Jiao, W.; Fang, Y.; Jian, P.; Shen, C.; Qin, Y.; Zhang, S.; et al. Improving rice nitrogen-use efficiency by modulating a novel monouniquitination machinery for optimal root plasticity response to nitrogen. Nat. Plants 2023, 9, 1902–1914. [Google Scholar] [CrossRef] [PubMed]
- Mounier, E.; Pervent, M.; Ljung, K.; Gojon, A.; Nacry, P. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 2014, 37, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubeš, M.; Pervent, M.; Léran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zažímalová, E.; et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cui, Y.; Yu, M.; Su, B.; Gong, W.; Baluska, F.; Komis, G.; Samaj, J.; Shan, X.; Lin, J. Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol. 2019, 181, 480–498. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Roudier, F.; Castaings, L.; Brehaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.C.; Ranathunge, K.; Melino, V.J.; Kuya, N.; Uga, Y.; Kronzucker, H.J. The intersection of nitrogen nutrition and water use in plants: New paths toward improved crop productivity. J. Exp. Bot. 2020, 71, 4452–4468. [Google Scholar] [CrossRef]
- Guo, S.; Zhou, Y.; Li, Y.; Gao, Y.; Shen, Q. Effects of different nitrogen forms and osmotic stress on water use efficiency of rice (Oryza sativa). Ann. Appl. Biol. 2008, 153, 127–134. [Google Scholar] [CrossRef]
- Waraich, E.A.; Ahmad, R.; Ashraf, M.Y.; Ahmad, M. Improving agricultural water use efficiency by nutrient management in crop plants. Acta Agric. Scand. Sect. B Soil Plant Sci. 2011, 61, 291–304. [Google Scholar] [CrossRef]
- Mao, Q.Z.; Watanabe, M.; Imori, M.; Kim, Y.S.; Kita, K.; Koike, T. Photosynthesis and nitrogen allocation in needles in the sun and shade crowns of hybrid larch saplings: Effect of nitrogen application. Photosynthetica 2012, 50, 422–428. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, C.; Huang, D.; Dong, Q.; Li, P.; Ma, F. Physiological evaluation of nitrogen use efficiency of different apple cultivars under various nitrogen and water supply conditions. J. Integr. Agric. 2020, 19, 709–720. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, J.; Guo, J.; Wang, Y.; Ji, H.; Chu, X.; Xiao, K.; Qi, X.; Hu, L.; Li, H.; et al. GmTDN1 improves wheat yields by inducing dual tolerance to both drought and low-N stress. Plant Biotechnol. J. 2022, 20, 1606–1621. [Google Scholar] [CrossRef] [PubMed]
- Araus, V.; Swift, J.; Alvarez, J.M.; Henry, A.; Coruzzi, G.M. A balancing act: How plants integrate nitrogen and water signals. J. Exp. Bot. 2020, 71, 4442–4451. [Google Scholar] [CrossRef] [PubMed]
- Cerda, A.; Alvarez, J.M. Insights into molecular links and transcription networks integrating drought stress and nitrogen signaling. New Phytol. 2024, 241, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Duan, X.; Zhao, X.; Ding, W.; Wang, Y.; Xiong, Y. Diverse nitrogen signals activate convergent ROP2-TOR signaling in Arabidopsis. Dev. Cell 2021, 56, 1283–1295.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 2018, 69, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Shanks, C.M.; Huang, J.; Cheng, C.Y.; Shih, H.J.S.; Brooks, M.D.; Alvarez, J.M.; Araus, V.; Swift, J.; Henry, A.; Coruzzi, G.M. Validation of a high-confidence regulatory network for gene-to-NUE phenotype infield-grown rice. Front. Plant Sci. 2022, 13, 1006044. [Google Scholar] [CrossRef]
- Friedrich, U.; von Oheimb, G.; Kriebitzsch, W.U.; Schlebelmann, K.; Weber, M.S.; Härdtle, W. Nitrogen deposition increases susceptibility to drought-experimental evidence with the perennial grass Moliniacaerulea (L.). Moench. Plant Soil 2012, 353, 59–71. [Google Scholar] [CrossRef]
- Dziedek, C.; Härdtle, W.; Oheimb, G.; Fichtner, A. Nitrogen addition enhances drought sensitivity of young deciduous tree species. Front. Plant Sci. 2016, 7, 1100. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.R.; Zhao, Q.; Yang, Y.Y.; Zhang, T.E.; Wang, X.F.; You, C.X.; Hao, Y.J. The apple 14-3-3 protein MdGRF11 interacts with the BTB protein MdBT2 to regulate nitrate deficiency-induced anthocyanin accumulation. Hort. Res. 2021, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Venail, J.; Mackay, S.; Bailey, P.C.; Schwinn, K.E.; Jameson, P.E.; Martin, C.R.; Davies, K.M. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011, 189, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Liu, J.; Lin, S.; Wang, J.; Lin, W.; Xu, W. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation. Plant Cell Rep. 2017, 36, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Xiao, W.; Mu, Q.; Li, D.; Chen, X.; Wu, H.; Li, L.; Peng, F. How does nitrate regulate plant senescence? Plant Physiol. Biochem. 2020, 157, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Utasee, S.; Jamjod, S.; Lordkaew, S.; Prom-U-Thai, C. Improve anthocyanin and zinc concentration in purple rice by nitrogen and zinc fertilizer application. Rice Sci. 2022, 29, 435–450. [Google Scholar] [CrossRef]
- Fredes, I.; Moreno, S.; Diaz, F.P.; Gutierrez, R.A. Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol. 2019, 47, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Gao, Y.; Han, M.; Liu, P.; Yang, C.; Shen, T.; Li, H. In vitro anthocyanin induction and metabolite analysis in Malus spectabilis leaves under low nitrogen conditions. Hortic. Plant J. 2020, 6, 284–292. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Yang, J.; Ma, C.; Zhang, Y.; Ge, T.; Qi, Z.; Kang, Y. Arabidopsis ROOT HAIR DEFECTIVE3 is involved in nitrogen starvation-induced anthocyanin accumulation. J. Integr. Plant Biol. 2015, 57, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Li, H.; An, X.; Song, L.; You, C.; Zhao, L.; Tian, Y.; Wang, X. MdMYB10 affects nitrogen uptake and reallocation by regulating the nitrate transporter MdNRT2.4-1 in red-fleshed apple. Hortic. Res. 2022, 9, uhac016. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, J.; Liu, X.; Su, L.; You, C.; Hao, Y. The nitrate-responsive protein MdBT2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiol. 2018, 178, 890–906. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Ren, Y.; Chen, A.; Yang, C.; Zheng, Q.; Chen, J.; Wang, D.; Li, Y.; Hu, S.; Xu, G. Plant nitrogen nutrition: The roles of arbuscular mycorrhizal fungi. J. Plant Physiol. 2022, 269, 153591. [Google Scholar] [CrossRef] [PubMed]
- Chalot, M.; Brun, A. Physiology of organic nitrogen acquisition by ectomycorrhizal fungi and ectomycorrhizas. FEMS Microbiol. Rev. 1998, 22, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Lozano, J.M.; Aroca, R.; Zamarreno, A.M.; Molina, S.; Andreo-Jimenez, B.; Porcel, R.; Garcia-Mina, J.M.; Ruyter-Spira, C.; Lopez-Raez, J.A. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 2016, 39, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, M.; Wang, Z.; Li, J.; Liu, K.; Huang, D. The role of arbuscular mycorrhizal symbiosis in plant abiotic stress. Front. Microbiol. 2024, 14, 1323881. [Google Scholar] [CrossRef] [PubMed]
- Zak, D.R.; Pellitier, P.T.; Argiroff, W.A.; Castillo, B.; James, T.Y.; Nave, L.E.; Averill, C.; Beidler, K.V.; Bhatnagar, J.; Blesh, J.; et al. Exploring the role of ectomycorrhizal fungi in soil carbon dynamics. New Phytol. 2019, 223, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, C.; Taniguchi, T.; Du, S.; Yamanaka, N.; Tateno, R. Soil nitrogen cycling is determined by the competition between mycorrhiza and ammonia-oxidizing prokaryotes. Ecology 2020, 101, e02963. [Google Scholar] [CrossRef] [PubMed]
- Bittebiere, A.K.; Vandenkoornhuyse, P.; Maluenda, E.; Gareil, A.; Dheilly, A.; Coudouel, S.; Bahin, M.; Mony, C. Past spatial structure of plant communities determines arbuscular mycorrhizal fungal community assembly. J. Ecol. 2019, 108, 546–560. [Google Scholar] [CrossRef]
- Tatsumi, C.; Hyodo, F.; Taniguchi, T.; Shi, W.Y.; Koba, K.; Fukushima, K.; Du, S.; Yamanaka, N.; Templer, P.; Tateno, R. Arbuscular mycorrhizal community in roots and nitrogen uptake patterns of understory trees beneath ectomycorrhizal and non-ectomycorrhizal overstory trees. Front. Plant Sci. 2021, 11, 583585. [Google Scholar] [CrossRef] [PubMed]
- Kobae, Y.; Tamura, Y.; Takai, S.; Banba, M.; Hata, S. Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol. 2010, 51, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tienda, J.; Valderas, A.; Camañes, G.; García-Agustín, P.; Ferrol, N. Kinetics of NH4+ uptake by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mycorrhiza 2012, 22, 485–491. [Google Scholar] [CrossRef]
- Calabrese, S.; Pérez-Tienda, J.; Ellerbeck, M.; Arnould, C.; Chatagnier, O.; Boller, T.; Schüßler, A.; Brachmann, A.; Wipf, D.; Ferrol, N.; et al. GintAMT3-a low-affinity ammonium transporter of the arbuscular mycorrhizal Rhizophagus irregularis. Front. Plant Sci. 2016, 7, 679. [Google Scholar] [CrossRef]
- Wang, S.; Chen, A.; Xie, K.; Yang, X.; Luo, Z.; Chen, J.; Zeng, D.; Ren, Y.; Yang, C.; Wang, L.; et al. Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 16649–16659. [Google Scholar] [CrossRef] [PubMed]
- Guether, M.; Volpe, V.; Balestrini, R.; Requena, N.; Wipf, D.; Bonfante, P. LjLHT1.2-a mycorrhiza-inducible plant amino acid transporter from Lotus japonicus. Biol. Fertil. Soils 2022, 47, 925–936. [Google Scholar] [CrossRef]
- Wang, Y.; Xuan, Y.; Wang, S.; Fan, D.; Wang, X.; Zheng, X. Genome-wide identification, characterization, and expression analysis of the ammonium transporter gene family in tea plants (Camellia sinensis L.). Physiol. Plant. 2022, 1, 174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, H.; Druzhinina, I.S.; Xie, X.; Wang, E.; Martin, F.; Yuan, Z. Phosphorus/nitrogen sensing and signaling in diverse root-fungus symbioses. Trends Microbiol. 2024, 32, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Saia, S.; Rappa, V.; Ruisi, P.; Abenavoli, M.R.; Sunseri, F.; Giambalvo, D.; Frenda, A.S.; Martinelli, F. Soil inoculation with symbiotic microorganisms promotes plant growth and nutrient transporter genes expression in durum wheat. Front. Plant Sci. 2015, 6, 815. [Google Scholar] [CrossRef] [PubMed]
- Hui, J.; An, X.; Li, Z.; Neuhäuser, B.; Ludewig, U.; Wu, X.; Schulze, W.X.; Chen, F.; Feng, G.; Lambers, H.; et al. The mycorrhiza-specific ammonium transporter ZmAMT3;1 mediates mycorrhiza-dependent nitrogen uptake in maize roots. Plant Cell 2022, 34, 4066–4087. [Google Scholar] [CrossRef] [PubMed]
- Lepetit, M.; Brouquisse, R. Control of the rhizobium–legume symbiosis by the plant nitrogen demand is tightly integrated at the whole plant level and requires inter-organ systemic signaling. Front. Plant Sci. 2023, 14, 1114840. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chen, R. Nitrogen and phosphorus signaling and transport during Legume–rhizobium symbiosis. Front. Plant Sci. 2021, 12, 683601. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, X.; Luo, Z.; Mysore, K.S.; Wen, J.; Xie, F. NIN interacts with NLPs to mediate NO3− inhibition of nodulation in Medicago truncatula. Nat. Plants 2018, 4, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Huang, Y.; Ren, Z.; Zhang, X.; Ren, J.; Su, J.; Zhang, C.; Tian, J.; Yu, Y.; Gao, G.F.; et al. Transfer cells mediate nitrate uptake to control root nodule symbiosis. Nat. Plants 2020, 6, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Valkov, V.T.; Sol, S.; Rogato, A.; Chiurazzi, M. The functional characterization of LjNRT2.4 indicates a novel, positive role of nitrate for an efficient nodule N2-ffxation activity. New Phytol. 2020, 228, 682–696. [Google Scholar] [CrossRef] [PubMed]
- Rogato, A.; D’Apuzzo, E.; Barbulova, A.; Omrane, S.; Stedel, C.; Simon-Rosin, U.; Katinakis, P.; Flemetakis, M.; Udvardi, M.; Chiurazzi, M. Tissue-speciffc down-regulation of LjAMT1;1 compromises nodule function and enhances nodulation in Lotus japonicus. Plant Mol. Biol. 2008, 68, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Hou, Y.; Xu, X. Optimized strategies for nitrogen fertilizer application in populus plantations in the context of climate change mitigation. For. Policy Econ. 2024, 159, 103139. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Li, S.; Li, J.; Huang, D. The Utilization and Roles of Nitrogen in Plants. Forests 2024, 15, 1191. https://doi.org/10.3390/f15071191
Wang Q, Li S, Li J, Huang D. The Utilization and Roles of Nitrogen in Plants. Forests. 2024; 15(7):1191. https://doi.org/10.3390/f15071191
Chicago/Turabian StyleWang, Qian, Shasha Li, Junrong Li, and Dong Huang. 2024. "The Utilization and Roles of Nitrogen in Plants" Forests 15, no. 7: 1191. https://doi.org/10.3390/f15071191
APA StyleWang, Q., Li, S., Li, J., & Huang, D. (2024). The Utilization and Roles of Nitrogen in Plants. Forests, 15(7), 1191. https://doi.org/10.3390/f15071191




