Soil pH and Nutrient Content Sustain Variability of Soil Bacterial Community Structure and Activity after Forest Clear-Cutting
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Soil Sampling
2.3. DNA Extraction and High-Throughput Sequencing
2.4. Environmental Analyses
2.5. Bioinformatic Analysis of Sequence Data
2.6. MicroResp Substrate-Induced Catabolic Activity Analysis
2.7. Data Preparation, Illustration, and Descriptive Statistics
3. Results
3.1. Site Characteristics
3.2. Bacterial Community Diversity and Composition
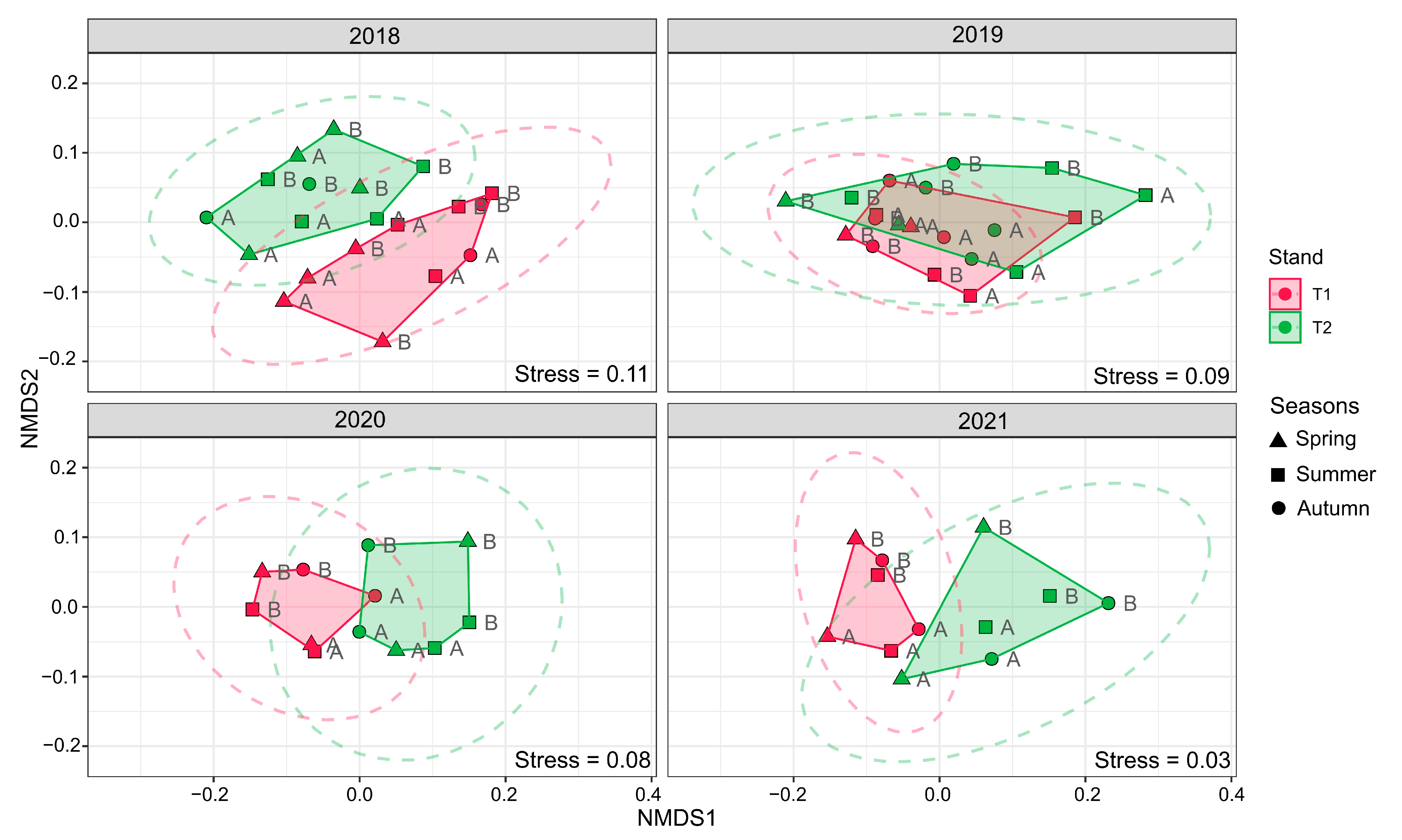
3.3. Bacterial Community Composition Shift
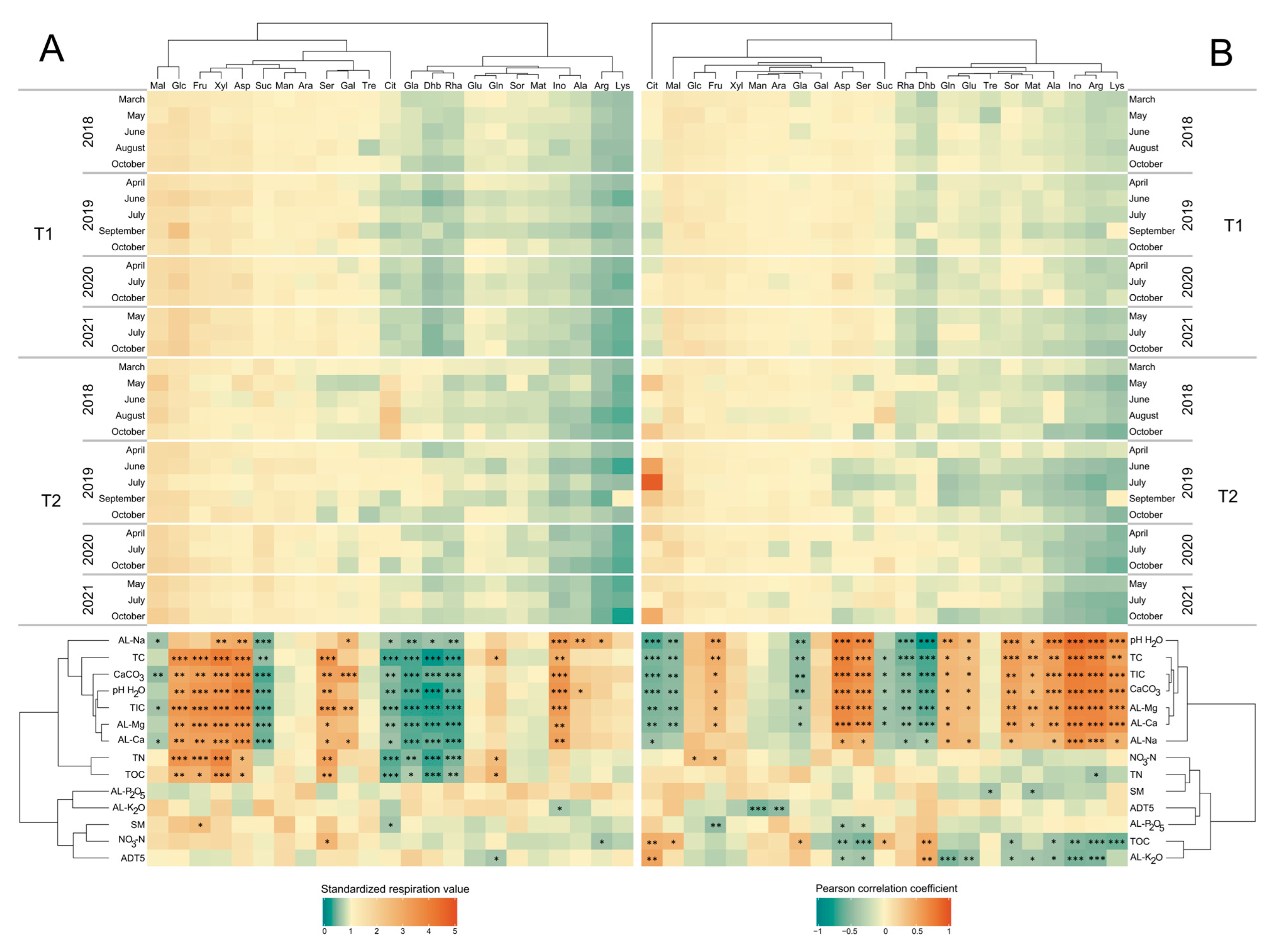
3.4. Microbial Respiration Activity
3.5. The Effects of Environmental Factors on Microbial Respiration
4. Discussion
4.1. Changes in Bacterial Community Diversity
4.2. Soil Bacterial Community Compositions Changes after Forest Harvest
4.3. The Effect of Forest Harvesting on Substrate-Induced Respiration
4.4. The Significance of Citrate in Bacterial Respiration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kembel, S.W.; Waters, I.; Shay, J.M. Short-Term Effects of Cut-to-Length versus Full-Tree Harvesting on Understorey Plant Communities and Understorey-Regeneration Associations in Manitoba Boreal Forests. For. Ecol. Manag. 2008, 255, 1848–1858. [Google Scholar] [CrossRef]
- Lencinas, M.V.; Martínez Pastur, G.; Gallo, E.; Cellini, J.M. Decreasing Negative Impacts of Harvesting over Insect Communities Using Variable Retention in Southern Patagonian Forests. J. Insect. Conserv. 2014, 18, 479–495. [Google Scholar] [CrossRef]
- Tozer, D.C.; Burke, D.M.; Nol, E.; Elliott, K.A. Short-Term Effects of Group-Selection Harvesting on Breeding Birds in a Northern Hardwood Forest. For. Ecol. Manag. 2010, 259, 1522–1529. [Google Scholar] [CrossRef]
- Closa, I.; Goicoechea, N. Seasonal Dynamics of the Physicochemical and Biological Properties of Soils in Naturally Regenerating, Unmanaged and Clear-Cut Beech Stands in Northern Spain. Eur. J. Soil Biol. 2010, 46, 190–199. [Google Scholar] [CrossRef]
- Nave, L.E.; Vance, E.D.; Swanston, C.W.; Curtis, P.S. Harvest Impacts on Soil Carbon Storage in Temperate Forests. For. Ecol. Manag. 2010, 259, 857–866. [Google Scholar] [CrossRef]
- Roy, M.-È.; Surget-Groba, Y.; Delagrange, S.; Rivest, D. Legacies of Forest Harvesting on Soil Properties along a Chronosequence in a Hardwood Temperate Forest. For. Ecol. Manag. 2021, 496, 119437. [Google Scholar] [CrossRef]
- Battigelli, J.P.; Spence, J.R.; Langor, D.W.; Berch, S.M. Short-Term Impact of Forest Soil Compaction and Organic Matter Removal on Soil Mesofauna Density and Oribatid Mite Diversity. Can. J. For. Res. 2004, 34, 1136–1149. [Google Scholar] [CrossRef]
- Elie, F.; Vincenot, L.; Berthe, T.; Quibel, E.; Zeller, B.; Saint-André, L.; Normand, M.; Chauvat, M.; Aubert, M. Soil Fauna as Bioindicators of Organic Matter Export in Temperate Forests. For. Ecol. Manag. 2018, 429, 549–557. [Google Scholar] [CrossRef]
- Rousseau, L.; Venier, L.; Aubin, I.; Gendreau-Berthiaume, B.; Moretti, M.; Salmon, S.; Handa, I.T. Woody Biomass Removal in Harvested Boreal Forest Leads to a Partial Functional Homogenization of Soil Mesofaunal Communities Relative to Unharvested Forest. Soil Biol. Biochem. 2019, 133, 129–136. [Google Scholar] [CrossRef]
- Hartmann, M.; Howes, C.G.; VanInsberghe, D.; Yu, H.; Bachar, D.; Christen, R.; Henrik Nilsson, R.; Hallam, S.J.; Mohn, W.W. Significant and Persistent Impact of Timber Harvesting on Soil Microbial Communities in Northern Coniferous Forests. ISME J. 2012, 6, 2199–2218. [Google Scholar] [CrossRef]
- Smenderovac, E.E.; Webster, K.; Caspersen, J.; Morris, D.; Hazlett, P.; Basiliko, N. Does Intensified Boreal Forest Harvesting Impact Soil Microbial Community Structure and Function? Can. J. For. Res. 2017, 47, 916–925. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Creamer, R.E.; Stone, D.; Berry, P.; Kuiper, I. Measuring Respiration Profiles of Soil Microbial Communities across Europe Using MicroRespTM Method. Appl. Soil Ecol. 2016, 97, 36–43. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-Biome Metagenomic Analyses of Soil Microbial Communities and Their Functional Attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S.; Wang, Z.; Zhang, Z.; Qin, H.; Wei, Z.; Feng, K.; Li, S.; Wu, Y.; Yin, H.; et al. Soil Microbiome Mediated Nutrients Decline during Forest Degradation Process. Soil Ecol. Lett. 2019, 1, 59–71. [Google Scholar] [CrossRef]
- Chodak, M.; Klimek, B.; Niklińska, M. Composition and Activity of Soil Microbial Communities in Different Types of Temperate Forests. Biol. Fertil. Soils 2016, 52, 1093–1104. [Google Scholar] [CrossRef]
- Dukunde, A.; Schneider, D.; Schmidt, M.; Veldkamp, E.; Daniel, R. Tree Species Shape Soil Bacterial Community Structure and Function in Temperate Deciduous Forests. Front. Microbiol. 2019, 10, 1519. [Google Scholar] [CrossRef]
- McCulley, R.L.; Archer, S.R.; Boutton, T.W.; Hons, F.M.; Zuberer, D.A. Soil Respiration and Nutrient Cycling in Wooded Communities Developing in Grassland. Ecology 2004, 85, 2804–2817. [Google Scholar] [CrossRef]
- Wu, S.-H.; Huang, B.-H.; Huang, C.-L.; Li, G.; Liao, P.-C. The Aboveground Vegetation Type and Underground Soil Property Mediate the Divergence of Soil Microbiomes and the Biological Interactions. Microb. Ecol. 2018, 75, 434–446. [Google Scholar] [CrossRef]
- Cardenas, E.; Kranabetter, J.M.; Hope, G.; Maas, K.R.; Hallam, S.; Mohn, W.W. Forest Harvesting Reduces the Soil Metagenomic Potential for Biomass Decomposition. ISME J. 2015, 9, 2465–2476. [Google Scholar] [CrossRef]
- Hynes, H.M.; Germida, J.J. Relationship between Ammonia Oxidizing Bacteria and Bioavailable Nitrogen in Harvested Forest Soils of Central Alberta. Soil Biol. Biochem 2012, 46, 18–25. [Google Scholar] [CrossRef]
- Mushinski, R.M.; Gentry, T.J.; Dorosky, R.J.; Boutton, T.W. Forest Harvest Intensity and Soil Depth Alter Inorganic Nitrogen Pool Sizes and Ammonia Oxidizer Community Composition. Soil Biol. Biochem 2017, 112, 216–227. [Google Scholar] [CrossRef]
- Danielson, R.E.; Mcginnis, M.L.; Holub, S.M.; Myrold, D.D. Soil Fungal and Prokaryotic Community Structure Exhibits Differential Short-Term Responses to Timber Harvest in the Pacific Northwest. Pedosphere 2020, 30, 109–125. [Google Scholar] [CrossRef]
- Kohout, P.; Charvátová, M.; Štursová, M.; Mašínová, T.; Tomšovský, M.; Baldrian, P. Clearcutting Alters Decomposition Processes and Initiates Complex Restructuring of Fungal Communities in Soil and Tree Roots. ISME J. 2018, 12, 692–703. [Google Scholar] [CrossRef]
- Churchland, C.; Bengtson, P.; Prescott, C.E.; Grayston, S.J. Dispersed Variable-Retention Harvesting Mitigates N Losses on Harvested Sites in Conjunction With Changes in Soil Microbial Community Structure. Front. For. Glob. Change 2021, 3, 609216. [Google Scholar] [CrossRef]
- Bååth, E.; Frostegård, Å.; Pennanen, T.; Fritze, H. Microbial Community Structure and pH Response in Relation to Soil Organic Matter Quality in Wood-Ash Fertilized, Clear-Cut or Burned Coniferous Forest Soils. Soil Biol. Biochem. 1995, 27, 229–240. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Cardenas, E.; Leung, H.; Szeitz, A.; Jensen, L.D.; Mohn, W.W. Long-Term Enrichment of Stress-Tolerant Cellulolytic Soil Populations Following Timber Harvesting Evidenced by Multi-Omic Stable Isotope Probing. Front. Microbiol. 2017, 8, 537. [Google Scholar] [CrossRef]
- Metzger, M.J.; Shkaruba, A.D.; Jongman, R.H.G.; Bunce, R.G.H. Descriptions of the European Environmental Zones and Strata; Alterra: Wageningen, The Netherlands, 2012; p. 154. [Google Scholar]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in Bacterial Communities along the 2000 Km Salinity Gradient of the Baltic Sea. ISME J 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Illumina 16S Metagenomic Sequencing Library Preparation. Available online: https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html (accessed on 13 February 2024).
- MSZ 21470-2:1981; Environmental Protection. Preparation of Soil Sample. Determination of Electrical Conduction, Humidity and pH. Hungarian Standards Institution: Budapest, Hungary, 1981. Available online: https://ugyintezes.mszt.hu/webaruhaz/szabvany-adatok?standard=79903 (accessed on 15 November 2018).
- MSZ-08-0206-2:1978; Evaluation of Some Chemical Properties of the Soil. Laboratory Tests. (pH Value, Phenolphtaleine Alkalinity Expressed in Soda, All Water Soluble Salts, Hydrolite (Yˇ1^-Value) and Exchanging Acidity (Yˇ2^-Value). Hungarian Standards Institution: Budapest, Hungary, 1978. Available online: https://ugyintezes.mszt.hu/webaruhaz/szabvanyadatok?standard=83417 (accessed on 15 November 2018).
- ISO 10694:1995; Soil Quality—Determination of Organic and Total Carbon after Dry Combustion (Elementary Analysis). International Organization for Standardization: Geneva, Switzerland, 1995. Available online: https://www.iso.org/standard/18782.html (accessed on 15 November 2018).
- ISO 13878:1998; Soil Quality—Determination of Total Nitrogen Content by Dry Combustion (“elemental Analysis”). International Organization for Standardization: Geneva, Switzerland, 1998. Available online: https://www.iso.org/standard/23117.html (accessed on 15 November 2018).
- MSZ 20135:1999; Determination of the Soluble Nutrient Element Content of the Soil. Hungarian Standards Institution: Budapest, Hungary, 1999. Available online: https://ugyintezes.mszt.hu/webaruhaz/szabvany-adatok?standard=97375 (accessed on 15 November 2018).
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the Wrinkles in the Rare Biosphere through Improved OTU Clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Prodan, A.; Tremaroli, V.; Brolin, H.; Zwinderman, A.H.; Nieuwdorp, M.; Levin, E. Comparing Bioinformatic Pipelines for Microbial 16S rRNA Amplicon Sequencing. PLoS ONE 2020, 15, e0227434. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Westcott, S.L.; Schloss, P.D. OptiClust, an Improved Method for Assigning Amplicon-Based Sequence Data to Operational Taxonomic Units. mSphere 2017, 2, e00073-17. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing 2022. Available online: https://www.R-project.org/ (accessed on 22 April 2022).
- Campbell, C.D.; Chapman, S.J.; Cameron, C.M.; Davidson, M.S.; Potts, J.M. A Rapid Microtiter Plate Method To Measure Carbon Dioxide Evolved from Carbon Substrate Amendments so as To Determine the Physiological Profiles of Soil Microbial Communities by Using Whole Soil. Appl. Environ. Microbiol. 2003, 69, 3593–3599. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D.; Software, P. PBC Dplyr: A Grammar of Data Manipulation 2023. Available online: https://dplyr.tidyverse.org/(accessed on 22 April 2023).
- Royston, J.P. Algorithm AS 181: The W Test for Normality. Appl. Stat. 1982, 31, 176. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Girden, E.R. ANOVA: Repeated Measures; SAGE Publications, Inc.: Thousand Oaks, CA, USA, 1992; ISBN 978-1-4129-8341-9. [Google Scholar]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Lenth, R.V.; Bolker, B.; Buerkner, P.; Giné-Vázquez, I.; Herve, M.; Jung, M.; Love, J.; Miguez, F.; Riebl, H.; Singmann, H. Emmeans: Estimated Marginal Means, Aka Least-Squares Means, R Package Version 1.8.6. 2023. Available online: https://CRAN.Rproject.org/package=emmeans (accessed on 15 October 2023).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Freedman, D.; Pisani, R.; Purves, R. Statistics, 4th ed.; W.W. Norton & Co: New York, NY, USA, 2007; ISBN 978-0-393-92972-0. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, R Package Version 2.6-2. 2022. Available online: https://CRAN.Rproject.org/package=vegan (accessed on 15 October 2023).
- Dray, S. Packfor: Forward Selection with Permutation (Canoco p.46), R Package Version 0.0-8/r136. 2016. Available online: https://RForge.R-project.org/projects/sedar/ (accessed on 15 October 2023).
- Kruskal, J.B. Nonmetric Multidimensional Scaling: A Numerical Method. Psychometrika 1964, 29, 115–129. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Use R! Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Brewster, R. Paint.Net 2023. Available online: https://getpaint.net/ (accessed on 15 December 2023).
- Lewandowski, T.E.; Forrester, J.A.; Mladenoff, D.J.; D’Amato, A.W.; Fassnacht, D.S.A.; Padley, E.; Martin, K.J. Do Biological Legacies Moderate the Effects of Forest Harvesting on Soil Microbial Community Composition and Soil Respiration. For. Ecol. Manag. 2019, 432, 298–308. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Kapusta, P.; Stanek, M.; Rola, K.; Zubek, S. Herbaceous Plant Species Support Soil Microbial Performance in Deciduous Temperate Forests. Sci. Total Environ. 2022, 810, 151313. [Google Scholar] [CrossRef] [PubMed]
- Henneron, L.; Cros, C.; Picon-Cochard, C.; Rahimian, V.; Fontaine, S. Plant Economic Strategies of Grassland Species Control Soil Carbon Dynamics through Rhizodeposition. J. Ecol. 2020, 108, 528–545. [Google Scholar] [CrossRef]
- Cornejo, N.S.; Becker, J.N.; Hemp, A.; Hertel, D. Effects of Land-Use Change and Disturbance on the Fine Root Biomass, Dynamics, Morphology, and Related C and N Fluxes to the Soil of Forest Ecosystems at Different Elevations at Mt. Kilimanjaro (Tanzania). Oecologia 2023, 201, 1089–1107. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, W.; Wu, M.; Xue, Y.; Ma, L.; Zhou, J. Effect of Aboveground Intervention on Fine Root Mass, Production, and Turnover Rate in a Chinese Cork Oak (Quercus variabilis Blume) Forest. Plant Soil 2013, 368, 201–214. [Google Scholar] [CrossRef]
- Martinović, T.; Kohout, P.; López-Mondéjar, R.; Algora Gallardo, C.; Starke, R.; Tomšovský, M.; Baldrian, P. Bacterial Community in Soil and Tree Roots of Picea abies Shows Little Response to Clearcutting. FEMS Microbiol. Ecol. 2022, 98, fiac118. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Juottonen, H.; Siivonen, P.; Lloret Quesada, C.; Tuomi, P.; Pulkkinen, P.; Yrjälä, K. Spatial Patterns of Microbial Diversity and Activity in an Aged Creosote-Contaminated Site. ISME J 2014, 8, 2131–2142. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Kuramae, E.E.; de Hollander, M.; Pijl, A.S.; van Veen, J.A.; Tsai, S.M. Acidobacterial Community Responses to Agricultural Management of Soybean in Amazon Forest Soils. FEMS Microbiol. Ecol. 2013, 83, 607–621. [Google Scholar] [CrossRef]
- Achat, D.L.; Deleuze, C.; Landmann, G.; Pousse, N.; Ranger, J.; Augusto, L. Quantifying Consequences of Removing Harvesting Residues on Forest Soils and Tree Growth—A Meta-Analysis. For. Ecol. Manag. 2015, 348, 124–141. [Google Scholar] [CrossRef]
- Gramss, G.; Voigt, K.-D.; Bergmann, H. Plant Availability and Leaching of (Heavy) Metals from Ammonium-, Calcium-, Carbohydrate-, and Citric Acid-Treated Uranium-Mine-Dump Soil. J. Plant Nutr. Soil Sci. 2004, 167, 417–427. [Google Scholar] [CrossRef]
- Evangelou, M.W.H.; Ebel, M.; Hommes, G.; Schaeffer, A. Biodegradation: The Reason for the Inefficiency of Small Organic Acids in Chelant-Assisted Phytoextraction. Water Air Soil Pollut. 2008, 195, 177–188. [Google Scholar] [CrossRef]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The Under-Recognized Dominance of Verrucomicrobia in Soil Bacterial Communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- da Rocha, U.N.; Andreote, F.D.; de Azevedo, J.L.; van Elsas, J.D.; van Overbeek, L.S. Cultivation of Hitherto-Uncultured Bacteria Belonging to the Verrucomicrobia Subdivision 1 from the Potato (Solanum tuberosum L.). Rhizosphere. J. Soils Sediments 2010, 10, 326–339. [Google Scholar] [CrossRef]
- Shen, C.; Ge, Y.; Yang, T.; Chu, H. Verrucomicrobial Elevational Distribution Was Strongly Influenced by Soil pH and Carbon/Nitrogen Ratio. J. Soils. Sediments 2017, 17, 2449–2456. [Google Scholar] [CrossRef]
- Ranjan, K.; Paula, F.S.; Mueller, R.C.; Jesus, E.d.C.; Cenciani, K.; Bohannan, B.J.M.; Nüsslein, K.; Rodrigues, J.L.M. Forest-to-Pasture Conversion Increases the Diversity of the Phylum Verrucomicrobia in Amazon Rainforest Soils. Front. Microbiol. 2015, 6, 779. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science 2013, 342, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Bai, X.; Zhou, Y.; Zhu, W.; Yin, Y. Variations of Soil Microbial Communities Accompanied by Different Vegetation Restoration in an Open-Cut Iron Mining Area. Sci. Total Environ. 2020, 704, 135243. [Google Scholar] [CrossRef]
- Lan, J.; Wang, S.; Wang, J.; Qi, X.; Long, Q.; Huang, M. The Shift of Soil Bacterial Community After Afforestation Influence Soil Organic Carbon and Aggregate Stability in Karst Region. Front. Microbiol. 2022, 13, 901126. [Google Scholar] [CrossRef]
- Kaiser, K.; Wemheuer, B.; Korolkow, V.; Wemheuer, F.; Nacke, H.; Schöning, I.; Schrumpf, M.; Daniel, R. Driving Forces of Soil Bacterial Community Structure, Diversity, and Function in Temperate Grasslands and Forests. Sci. Rep. 2016, 6, 33696. [Google Scholar] [CrossRef] [PubMed]
- Plassart, P.; Prévost-Bouré, N.C.; Uroz, S.; Dequiedt, S.; Stone, D.; Creamer, R.; Griffiths, R.I.; Bailey, M.J.; Ranjard, L.; Lemanceau, P. Soil Parameters, Land Use, and Geographical Distance Drive Soil Bacterial Communities along a European Transect. Sci. Rep. 2019, 9, 605. [Google Scholar] [CrossRef]
- Houston, A.P.C.; Visser, S.; Lautenschlager, R.A. Microbial Processes and Fungal Community Structure in Soils from Clear-Cut and Unharvested Areas of Two Mixedwood Forests. Can. J. Bot. 1998, 76, 630–640. [Google Scholar] [CrossRef]
- Johnson, D.; Booth, R.E.; Whiteley, A.S.; Bailey, M.J.; Read, D.J.; Grime, J.P.; Leake, J.R. Plant Community Composition Affects the Biomass, Activity and Diversity of Microorganisms in Limestone Grassland Soil. Eur. J. Soil Sci. 2003, 54, 671–678. [Google Scholar] [CrossRef]
- Orwin, K.H.; Buckland, S.M.; Johnson, D.; Turner, B.L.; Smart, S.; Oakley, S.; Bardgett, R.D. Linkages of Plant Traits to Soil Properties and the Functioning of Temperate Grassland. J. Ecol. 2010, 98, 1074–1083. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Kapusta, P.; Błońska, A.; Kompała-Bąba, A.; Woźniak, G. Effects of Calamagrostis Epigejos, Chamaenerion Palustre and Tussilago Farfara on Nutrient Availability and Microbial Activity in the Surface Layer of Spoil Heaps after Hard Coal Mining. Ecol. Eng. 2015, 83, 328–337. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Kapusta, P.; Stanek, M.; Rożek, K.; Rola, K.; Zubek, S. Herbaceous Plant Species and Their Combinations Positively Affect Soil Microorganisms and Processes and Modify Soil Physicochemical Properties in a Mesocosm Experiment. For. Ecol. Manag. 2023, 532, 120826. [Google Scholar] [CrossRef]
- Roberts, M.R.; Zhu, L. Early Response of the Herbaceous Layer to Harvesting in a Mixed Coniferous–Deciduous Forest in New Brunswick, Canada. For. Ecol. Manag. 2002, 155, 17–31. [Google Scholar] [CrossRef]
- de Graaf, M.; Roberts, M.R. Short-Term Response of the Herbaceous Layer within Leave Patches after Harvest. For. Ecol. Manag. 2009, 257, 1014–1025. [Google Scholar] [CrossRef]
- Nguyen, C. Rhizodeposition of Organic C by Plant: Mechanisms and Controls. In Sustainable Agriculture; Lichtfouse, E., Navarrete, M., Debaeke, P., Véronique, S., Alberola, C., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 97–123. ISBN 978-90-481-2666-8. [Google Scholar]
- Jones, D.L.; Hodge, A.; Kuzyakov, Y. Plant and Mycorrhizal Regulation of Rhizodeposition. New Phytol. 2004, 163, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are Root Exudates More Important than Other Sources of Rhizodeposits in Structuring Rhizosphere Bacterial Communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Struecker, J.; Joergensen, R.G. Microorganisms and Their Substrate Utilization Patterns in Topsoil and Subsoil Layers of Two Silt Loams, Differing in Soil Organic C Accumulation Due to Colluvial Processes. Soil Biol. Biochem. 2015, 91, 310–317. [Google Scholar] [CrossRef]
- Gunina, A.; Dippold, M.A.; Glaser, B.; Kuzyakov, Y. Fate of Low Molecular Weight Organic Substances in an Arable Soil: From Microbial Uptake to Utilisation and Stabilisation. Soil Biol. Biochem. 2014, 77, 304–313. [Google Scholar] [CrossRef]
- Kaneda, S.; Krištůfek, V.; Baldrian, P.; Malý, S.; Frouz, J. Changes in Functional Response of Soil Microbial Community along Chronosequence of Spontaneous Succession on Post Mining Forest Sites Evaluated by Biolog and SIR Methods. Forests 2019, 10, 1005. [Google Scholar] [CrossRef]
- Bongiorno, G.; Bünemann, E.K.; Brussaard, L.; Mäder, P.; Oguejiofor, C.U.; de Goede, R.G.M. Soil Management Intensity Shifts Microbial Catabolic Profiles across a Range of European Long-Term Field Experiments. Appl. Soil Ecol. 2020, 154, 103596. [Google Scholar] [CrossRef]
- Brockett, B.F.T.; Prescott, C.E.; Grayston, S.J. Soil Moisture Is the Major Factor Influencing Microbial Community Structure and Enzyme Activities across Seven Biogeoclimatic Zones in Western Canada. Soil Biol. Biochem. 2012, 44, 9–20. [Google Scholar] [CrossRef]
- Kania, A.; Langlade, N.; Martinoia, E.; Neumann, G. Phosphorus Deficiency-Induced Modifications in Citrate Catabolism and in Cytosolic pH as Related to Citrate Exudation in Cluster Roots of White Lupin. Plant Soil 2003, 248, 117–127. [Google Scholar] [CrossRef]
- Menezes-Blackburn, D.; Paredes, C.; Zhang, H.; Giles, C.D.; Darch, T.; Stutter, M.; George, T.S.; Shand, C.; Lumsdon, D.; Cooper, P.; et al. Organic Acids Regulation of Chemical–Microbial Phosphorus Transformations in Soils. Environ. Sci. Technol. 2016, 50, 11521–11531. [Google Scholar] [CrossRef]
- Oral, A.; Uygur, V. Effects of Low-Molecular-Mass Organic Acids on P Nutrition and Some Plant Properties of Hordeum vulgare. J. Plant Nutr. 2018, 41, 1482–1490. [Google Scholar] [CrossRef]
- Akram, M. Citric Acid Cycle and Role of Its Intermediates in Metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Dimroth, P. Molecular Basis for Bacterial Growth on Citrate or Malonate. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, R.J.; Bériault, R.; Lemire, J.; Singh, R.; Chénier, D.R.; Hamel, R.D.; Appanna, V.D. The Tricarboxylic Acid Cycle, an Ancient Metabolic Network with a Novel Twist. PLoS ONE 2007, 2, e690. [Google Scholar] [CrossRef] [PubMed]
- Meylan, S.; Porter, C.B.M.; Yang, J.H.; Belenky, P.; Gutierrez, A.; Lobritz, M.A.; Park, J.; Kim, S.H.; Moskowitz, S.M.; Collins, J.J. Carbon Sources Tune Antibiotic Susceptibility in Pseudomonas Aeruginosa via Tricarboxylic Acid Cycle Control. Cell Chem. Biol. 2017, 24, 195–206. [Google Scholar] [CrossRef]
- Ramond, E.; Gesbert, G.; Rigard, M.; Dairou, J.; Dupuis, M.; Dubail, I.; Meibom, K.; Henry, T.; Barel, M.; Charbit, A. Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Francisella Phagosomal Escape. PLoS Pathog. 2014, 10, e1003893. [Google Scholar] [CrossRef]



| Layer | Stand | Year | Soil Moisture | CaCO3 | TN | TC | TOC | TIC | |
|---|---|---|---|---|---|---|---|---|---|
| % | % | % | % | % | % | ||||
| A | T1 | 2018 | 17.10 ± 6.67 a | 7.17 ± 0.09 b | 5.41 ± 1.15 b | 0.20 ± 0.01 ab | 3.65 ± 0.19 bc | 2.44 ± 0.13 ab | 1.17 ± 0.19 b |
| 2019 | 17.0 ± 2.59 a | 7.31 ± 0.10 b | 7.02 ± 1.41 b | 0.26 ± 0.03 bc | 4.08 ± 0.28 cd | 2.80 ± 0.36 abc | 1.28 ± 0.22 b | ||
| 2020 | 16.1 ± 3.29 a | 7.24 ± 0.04 b | 5.30 ± 2.10 b | 0.27 ± 0.01 bc | 4.45 ± 0.58 cd | 2.98 ± 0.34 bc | 1.47 ± 0.42 b | ||
| 2021 | 23.1 ± 1.84 a | 7.59 ± 0.15 b | 4.92 ± 0.42 b | 0.31 ± 0.03 c | 4.68 ± 0.15 d | 3.26 ± 0.07 c | 1.42 ± 0.08 b | ||
| T2 | 2018 | 15.5 ± 5.74 a | 5.82 ± 0.6 a | 0 ± 0 a | 0.16 ± 0.02 a | 2.38 ± 0.25 a | 2.22 ± 0.20 a | 0.17 ± 0.06 a | |
| 2019 | 18.5 ± 3.90 a | 5.76 ± 0.87 a | 0.22 ± 0.49 a | 0.22 ± 0.05 ab | 2.90 ± 0.53 ab | 2.58 ± 0.41 abc | 0.32 ± 0.16 a | ||
| 2020 | 18.6 ± 7.08 a | 5.50 ± 0.61 a | 0 ± 0 a | 0.23 ± 0.06 abc | 3.19 ± 0.28 ab | 2.86 ± 0.21 abc | 0.33 ± 0.07 a | ||
| 2021 | 20.1 ± 3.63 a | 6.44 ± 0.38 ab | 0.82 ± 1.42 a | 0.25 ± 0.05 bc | 3.53 ± 0.17 bc | 3.02 ± 0.25 bc | 0.51 ± 0.19 a | ||
| Layer | Stand | Year | AL-P2O5 | AL-K2O | AL-Na | AL-Mg | AL-Ca | NO3-N | |
| mg 100 g−1 | mg 100 g−1 | mg kg−1 | mg g−1 | mg g−1 | mg kg−1 | ||||
| A | T1 | 2018 | 10.4 ± 3.21 a | 21.8 ± 1.86 a | 80.4 ± 21.7 c | 1.05 ± 0.53 abc | 25.0 ± 19.7 bcd | 2.81 ± 2.53 a | |
| 2019 | 10.0 ± 2.96 a | 29.2 ± 2.22 bc | 74.5 ± 15.1 bc | 1.20 ± 0.16 c | 30.9 ± 5.89 d | 0.92 ± 0.44 a | |||
| 2020 | 6.74 ± 4.96 a | 29.2 ± 3.42 abc | 61.2 ± 7.04 abc | 1.27 ± 0.07 c | 30.6 ± 14.2 cd | 1.55 ± 2.02 a | |||
| 2021 | 7.69 ± 0.98 a | 23.4 ± 0.69 ab | 60.6 ± 1.24 abc | 1.17 ± 0.03 bc | 25.7 ± 3.47 abcd | 4.41 ± 2.18 a | |||
| T2 | 2018 | 8.74 ± 2.18 a | 22.9 ± 2.32 ab | 42.6 ± 9.10 a | 0.51 ± 0.06 a | 2.43 ± 0.741 a | 1.29 ± 1.94 a | ||
| 2019 | 12.7 ± 3.17 a | 35.5 ± 5.03 c | 49.0 ± 0.59 ab | 0.56 ± 0.15 ab | 3.26 ± 2.70 a | 1.18 ± 1.48 a | |||
| 2020 | 8.64 ± 4.84 a | 35.6 ± 3.67 c | 50.3 ± 3.09 abc | 0.55 ± 0.11 ab | 2.28 ± 0.53 ab | 0.78 ± 1.35 a | |||
| 2021 | 8.86 ± 0.76 a | 32.7 ± 0.31 c | 49.3 ± 8.53 abc | 0.71 ± 0.18 abc | 5.88 ± 5.61 abc | 4.59 ± 2.06 a | |||
| Layer | Stand | Year | Soil Moisture | CaCO3 | TN | TC | TOC | TIC | |
| % | % | % | % | % | % | ||||
| B | T1 | 2018 | 14.80 ± 5.69 a | 7.54 ± 0.16 b | 10.6 ± 2.39 b | 0.13 ± 0.01 a | 3.45 ± 0.34 b | 1.62 ± 0.14 ab | 1.81 ± 0.44 b |
| 2019 | 15.1 ± 3.22 a | 7.73 ± 0.08 b | 9.83 ± 2.06 b | 0.14 ± 0.02 a | 3.15 ± 0.12 b | 1.42 ± 0.14 a | 1.73 ± 0.24 b | ||
| 2020 | 12.4 ± 1.21 a | 7.60 ± 0.09 b | 8.70 ± 0.57 b | 0.15 ± 0.02 a | 3.30 ± 0.18 b | 1.49 ± 0.15 ab | 1.80 ± 0.22 b | ||
| 2021 | 17.1 ± 1.07 a | 8.03 ± 0.18 b | 7.80 ± 1.47 b | 0.17 ± 0.02 a | 3.16 ± 0.14 b | 1.58 ± 0.19 ab | 1.58 ± 0.19 b | ||
| T2 | 2018 | 14.00 ± 4.48 a | 6.37 ± 0.41 a | 0 ± 0 a | 0.15 ± 0.01 a | 2.22 ± 0.15 a | 2.06 ± 0.08 c | 0.20 ± 0.08 a | |
| 2019 | 17.9 ± 3.76 a | 6.03 ± 0.96 a | 1.41 ± 3.15 a | 0.16 ± 0.01 a | 2.26 ± 0.50 a | 1.80 ± 0.28 bc | 0.47 ± 0.63 a | ||
| 2020 | 17.5 ± 2.81 a | 5.33 ± 0.28 a | 0 ± 0 a | 0.16 ± 0.05 a | 2.02 ± 0.13 a | 1.84 ± 0.11 bc | 0.18 ± 0.02 a | ||
| 2021 | 17.2 ± 1.54 a | 5.86 ± 0.48 a | 0 ± 0 a | 0.16 ± 0.01 a | 2.00 ± 0.14 a | 1.81 ± 0.11 abc | 0.19 ± 0.03 a | ||
| Layer | Stand | Year | AL-P2O5 | AL-K2O | AL-Na | AL-Mg | AL-Ca | NO3-N | |
| mg 100 g−1 | mg 100 g−1 | mg kg−1 | mg g−1 | mg g−1 | mg kg−1 | ||||
| B | T1 | 2018 | 8.72 ± 3.30 a | 16.6 ± 2.15 a | 83.2 ± 22.0 bc | 1.22 ± 0.41 bc | 34.5 ± 20.1 cd | 0.75 ± 1.30 a | |
| 2019 | 7.64 ± 3.39 a | 21.3 ± 2.05 b | 85.5 ± 10.9 c | 1.35 ± 0.16 c | 47.6 ± 7.31 d | 1.70 ± 1.82 a | |||
| 2020 | 3.91 ± 4.96 a | 23.0 ± 2.12 bc | 65.8 ± 17.4 abc | 1.49 ± 0.08 c | 43.3 ± 14.3 d | 0.64 ± 0.67 a | |||
| 2021 | 4.26 ± 0.80 a | 19.1 ± 1.79 ab | 69.0 ± 4.61 abc | 1.24 ± 0.01 bc | 37.5 ± 4.55 bcd | 2.75 ± 1.61 a | |||
| T2 | 2018 | 8.41 ± 3.47 a | 22.1 ± 1.19 bc | 50.8 ± 7.16 a | 0.48 ± 0.04 a | 3.09 ± 0.81 a | 0.09 ± 0.19 a | ||
| 2019 | 10.4 ± 2.75 a | 25.8 ± 2.83 c | 55.8 ± 12.7 ab | 0.66 ± 0.45 ab | 10.8 ± 19.0 abc | 0.81 ± 1.38 a | |||
| 2020 | 5.37 ± 4.41 a | 23.9 ± 1.81 bc | 56.5 ± 4.75 abc | 0.47 ± 0.06 a | 2.28 ± 0.12 ab | 0.17 ± 0.23 a | |||
| 2021 | 5.62 ± 1.76 a | 22.2 ± 0.28 bc | 47.0 ± 1.99 a | 0.49 ± 0.01 a | 2.33 ± 0.12 ab | 2.51 ± 0.76 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bereczki, K.; Benke, A.; Tóth, E.G.; Megyes, M.; Korponai, K.; Szili-Kovács, T.; Illés, G.; Lados, B.B.; Márialigeti, K. Soil pH and Nutrient Content Sustain Variability of Soil Bacterial Community Structure and Activity after Forest Clear-Cutting. Forests 2024, 15, 1284. https://doi.org/10.3390/f15081284
Bereczki K, Benke A, Tóth EG, Megyes M, Korponai K, Szili-Kovács T, Illés G, Lados BB, Márialigeti K. Soil pH and Nutrient Content Sustain Variability of Soil Bacterial Community Structure and Activity after Forest Clear-Cutting. Forests. 2024; 15(8):1284. https://doi.org/10.3390/f15081284
Chicago/Turabian StyleBereczki, Katalin, Attila Benke, Endre György Tóth, Melinda Megyes, Kristóf Korponai, Tibor Szili-Kovács, Gábor Illés, Botond Boldizsár Lados, and Károly Márialigeti. 2024. "Soil pH and Nutrient Content Sustain Variability of Soil Bacterial Community Structure and Activity after Forest Clear-Cutting" Forests 15, no. 8: 1284. https://doi.org/10.3390/f15081284






