Increased Saprotrophic Activity and Phosphate Leaching Following Forest Soil Decomposition without Root Access
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Soil Decomposition Experiment—Sampling, Analyses and Design
2.3. Soil Analyses
2.3.1. Soil Chemistry
2.3.2. Soil Microbial Biomass and Extractable Organic Carbon
2.3.3. Extracellular Hydrolytic Enzyme Activity
2.3.4. DNA Extraction and Quantification
2.3.5. Microbial Community Composition Analysis and Lifestyle Assignment
2.4. Statistical Analyses
3. Results
3.1. Initial Soil Composition and Biochemistry
3.2. Initial Soil Microbiome Composition
3.3. Soil Response to Incubation without Root Access
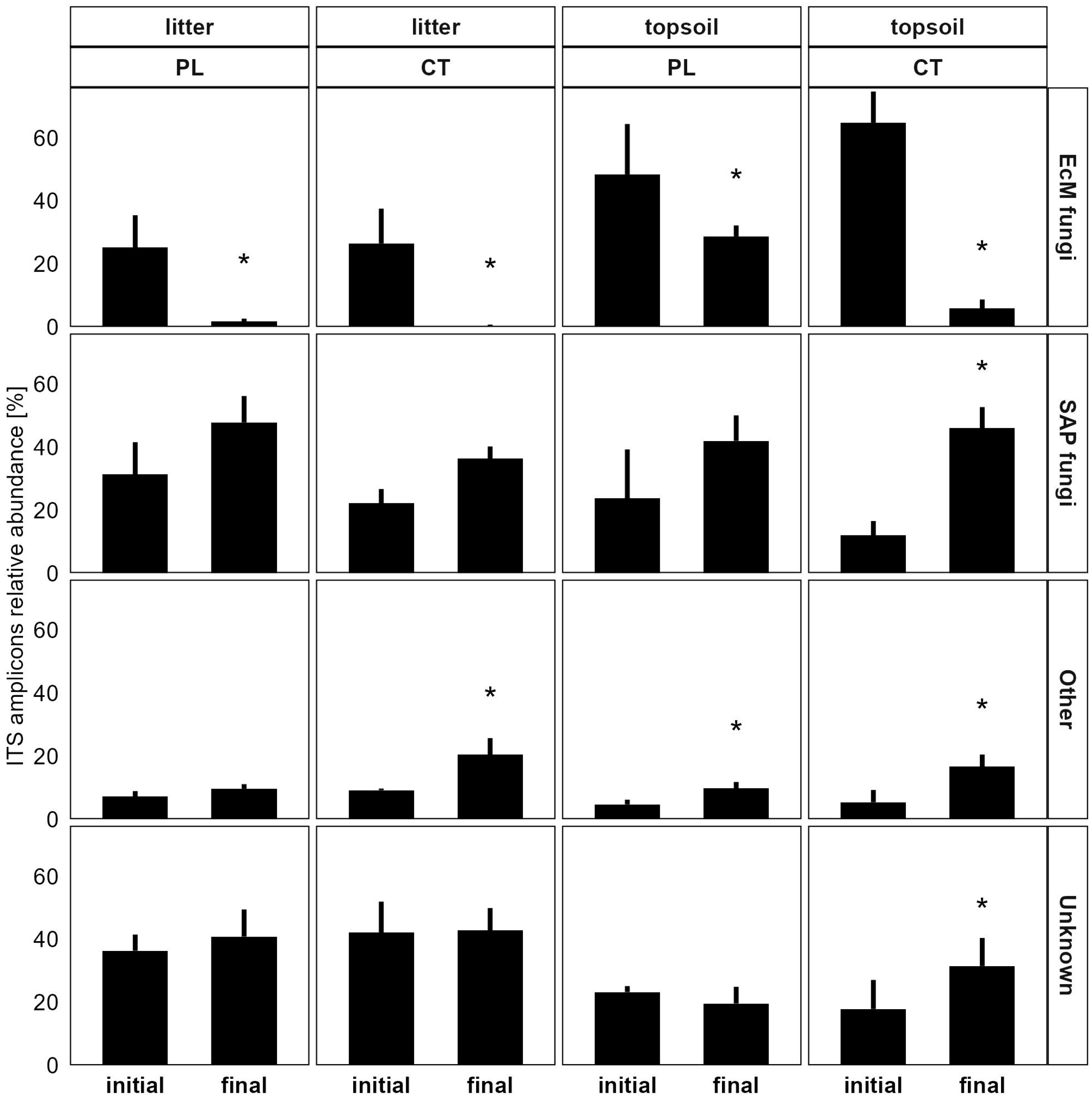
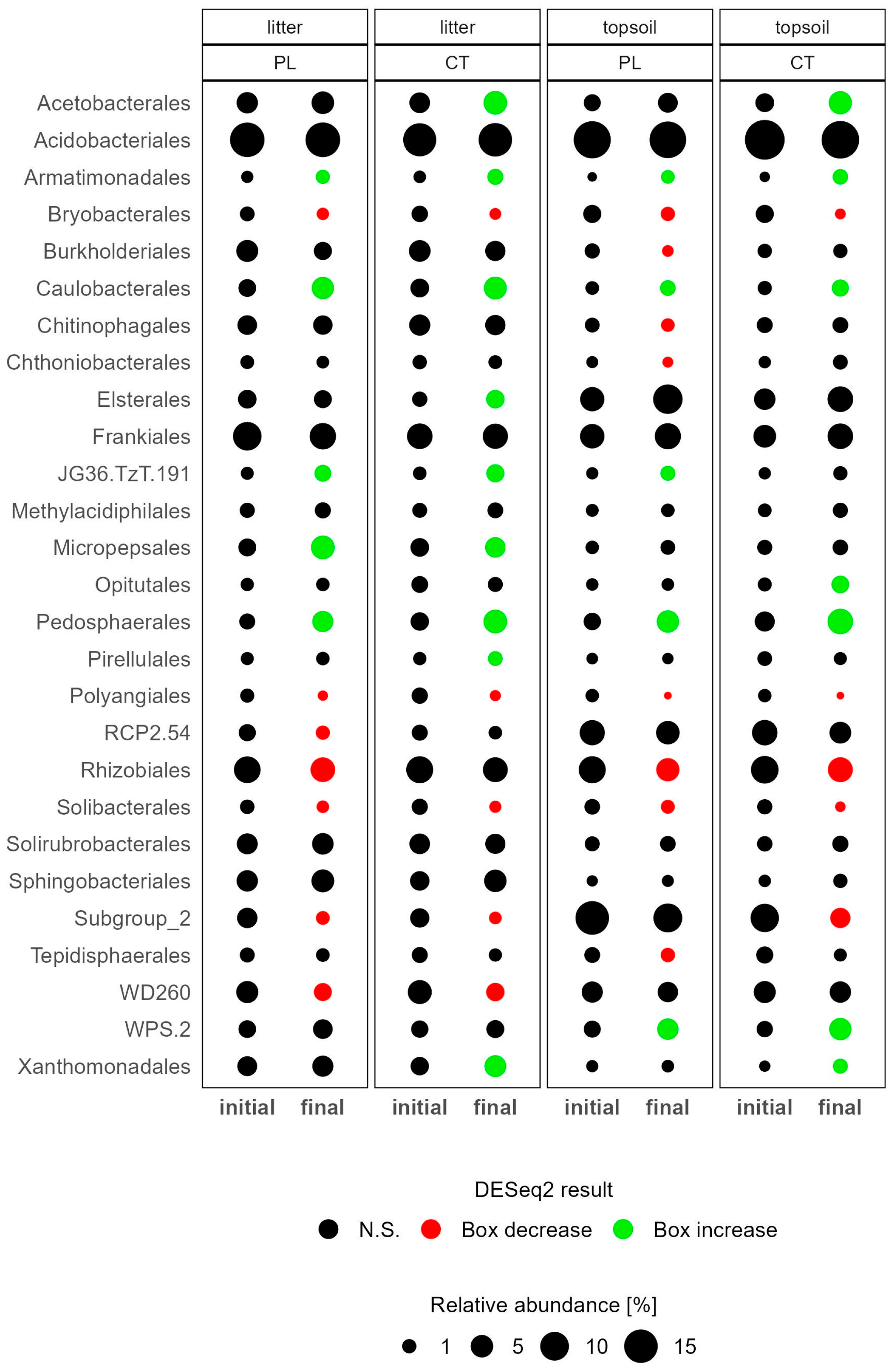
3.3.1. Soil Microbiome Response
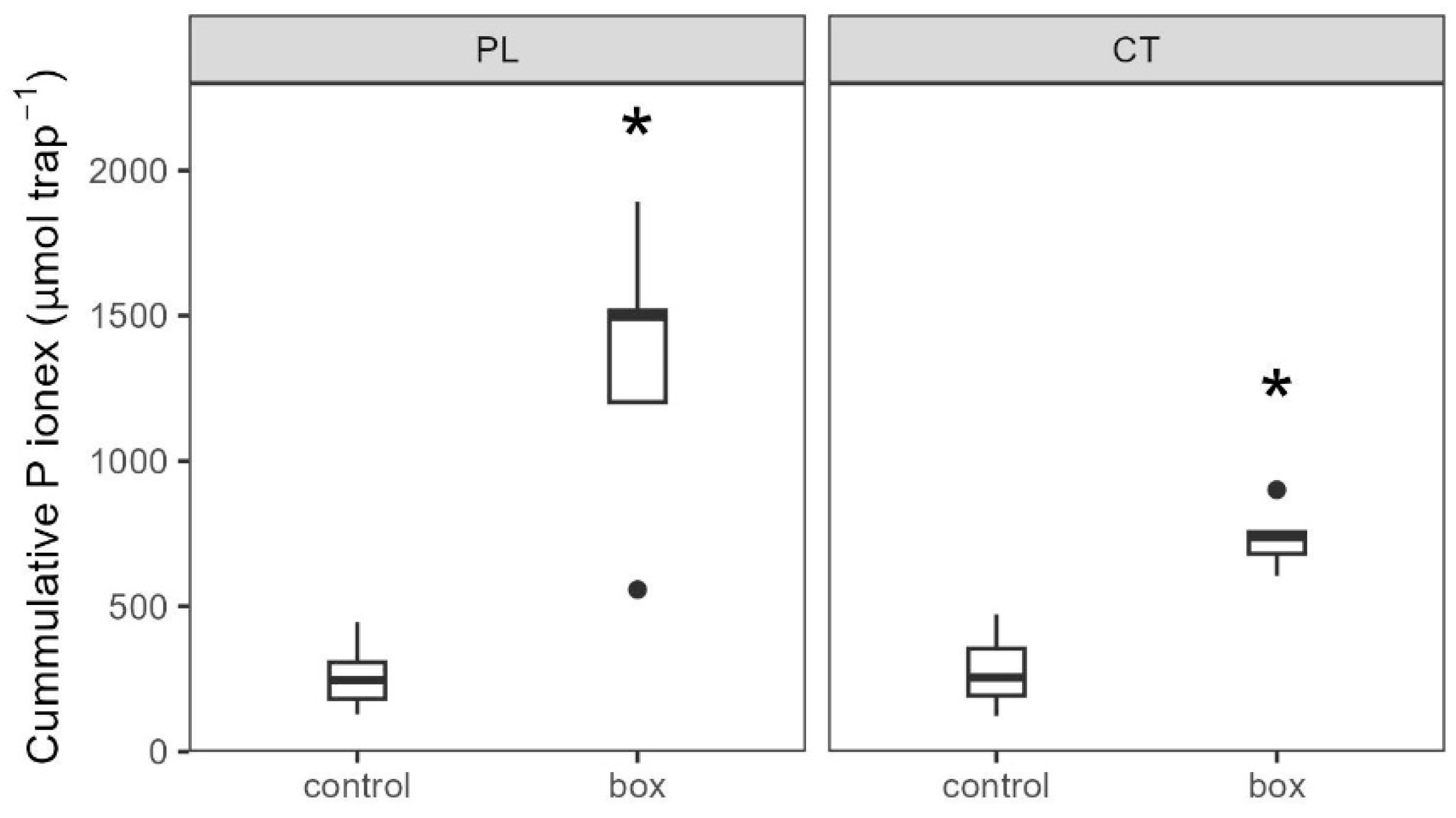
3.3.2. Soil Biochemistry Changes
4. Discussion
4.1. Microbiome Changes
4.2. Intensification of P Cycling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bödeker, I.T.M.; Lindahl, B.D.; Olson, Å.; Clemmensen, K.E. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 2016, 30, 1967–1978. [Google Scholar] [CrossRef]
- Hobbie, E.A. Carbon allocation to ectomycorrhizal fungi correlates with belowground allocation in culture studies. Ecology 2006, 87, 563–569. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Tunlid, A. Ectomycorrhizal fungi—Potential organic matter decomposers, yet not saprotrophs. New Phytol. 2015, 205, 1443–1447. [Google Scholar] [CrossRef]
- Zak, D.R.; Pellitier, P.T.; Argiroff, W.A.; Castillo, B.; James, T.Y.; Nave, L.E.; Averill, C.; Beidler, K.V.; Bhatnagar, J.; Blesh, J.; et al. Exploring the role of ectomycorrhizal fungi in soil carbon dynamics. New Phytol. 2019, 223, 33–39. [Google Scholar] [CrossRef]
- Averill, C.; Turner, B.L.; Finzi, A.C. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 2014, 505, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.R.; Wan, J. Resource-ratio theory predicts mycorrhizal control of litter decomposition. New Phytol. 2019, 223, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.W.; Kennedy, P.G. Revisiting the ‘Gadgil effect’: Do interguild fungal interactions control carbon cycling in forest soils? New Phytol. 2016, 209, 1382–1394. [Google Scholar] [CrossRef]
- LeBauer, D.S.; Treseder, K.K. Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology 2008, 89, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Gadgil, R.L.; Gadgil, P.D. Mycorrhiza and Litter Decomposition. Nature 1971, 233, 133. [Google Scholar] [CrossRef]
- Boberg, J.B.; Finlay, R.D.; Stenlid, J.; Ekblad, A.; Lindahl, B.D. Nitrogen and Carbon Reallocation in Fungal Mycelia during Decomposition of Boreal Forest Litter. PLoS ONE 2014, 9, e92897. [Google Scholar] [CrossRef]
- Averill, C.; Hawkes, C.V. Ectomycorrhizal fungi slow soil carbon cycling. Ecol. Lett. 2016, 19, 937–947. [Google Scholar] [CrossRef]
- Gadgil, P.D. Suppression of Litter Decomposition by Mycorrhizal Roots of Pinus radiata/Ruth L. Gadgil and P.D. Gadgil; New Zealand Forest Service: Wellington, New Zealand, 1975. [Google Scholar]
- Tedersoo, L.; Bahram, M. Mycorrhizal types differ in ecophysiology and alter plant nutrition and soil processes. Biol. Rev. 2019, 94, 1857–1880. [Google Scholar] [CrossRef]
- Högberg, M.N.; Högberg, P. Extramatrical ectomycorrhizal mycelium contributes one-third of microbial biomass and produces, together with associated roots, half the dissolved organic carbon in a forest soil. New Phytol. 2002, 154, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Phillips, L.A.; Ward, V.; Jones, M.D. Ectomycorrhizal fungi contribute to soil organic matter cycling in sub-boreal forests. Isme J. 2014, 8, 699–713. [Google Scholar] [CrossRef]
- Čada, V.; Morrissey, R.C.; Michalová, Z.; Bače, R.; Janda, P.; Svoboda, M. Frequent severe natural disturbances and non-equilibrium landscape dynamics shaped the mountain spruce forest in central Europe. For. Ecol. Manag. 2016, 363, 169–178. [Google Scholar] [CrossRef]
- Štursová, M.; Šnajdr, J.; Cajthaml, T.; Bárta, J.; Šantrůčková, H.; Baldrian, P. When the forest dies: The response of forest soil fungi to a bark beetle-induced tree dieback. ISME J. 2014, 8, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.M.; Turner, M.G. Changes to the N cycle following bark beetle outbreaks in two contrasting conifer forest types. Oecologia 2012, 170, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.M.; Turner, M.G.; Simard, M. Nitrogen cycling following mountain pine beetle disturbance in lodgepole pine forests of Greater Yellowstone. For. Ecol. Manag. 2011, 261, 1077–1089. [Google Scholar] [CrossRef]
- Huber, C. Long lasting nitrate leaching after bark beetle attack in the highlands of the Bavarian Forest National Park. J Environ. Qual. 2005, 34, 1772–1779. [Google Scholar] [CrossRef]
- Cigan, P.W.; Karst, J.; Cahill, J.F.; Sywenky, A.N.; Pec, G.J.; Erbilgin, N. Influence of bark beetle outbreaks on nutrient cycling in native pine stands in western Canada. Plant Soil 2015, 390, 29–47. [Google Scholar] [CrossRef]
- Norton, U.; Ewers, B.E.; Borkhuu, B.; Brown, N.R.; Pendall, E. Soil Nitrogen Five Years after Bark Beetle Infestation in Lodgepole Pine Forests. Soil Sci. Soc. Am. J. 2015, 79, 282–293. [Google Scholar] [CrossRef]
- Kaňa, J.; Tahovská, K.; Kopáček, J. Response of soil chemistry to forest dieback after bark beetle infestation. Biogeochemistry 2013, 113, 369–383. [Google Scholar] [CrossRef]
- Du, E.; van Doorn, M.; de Vries, W. Spatially divergent trends of nitrogen versus phosphorus limitation across European forests. Sci. Total Environ. 2021, 771, 145391. [Google Scholar] [CrossRef] [PubMed]
- Clarholm, M.; Skyllberg, U.; Rosling, A. Organic acid induced release of nutrients from metal-stabilized soil organic matter—The unbutton model. Soil Biol. Biochem. 2015, 84, 168–176. [Google Scholar] [CrossRef]
- Jansa, J.; Finlay, R.; Wallander, H.; Smith, F.A.; Smith, S.E. Role of Mycorrhizal Symbioses in Phosphorus Cycling. In Phosphorus in Action: Biological Processes in Soil Phosphorus Cycling; Bünemann, E., Oberson, A., Frossard, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 137–168. [Google Scholar]
- Kopáček, J.; Čapek, P.; Choma, M.; Cudlín, P.; Kaňa, J.; Kopáček, M.; Porcal, P.; Šantrůčková, H.; Tahovská, K.; Turek, J. Long-term changes in soil composition in unmanaged central European mountain spruce forests after decreased acidic deposition and a bark beetle outbreak. Catena 2023, 222, 106839. [Google Scholar] [CrossRef]
- Kopáček, J.; Hejzlar, J.; Kaňa, J.; Porcal, P.; Turek, J. The sensitivity of water chemistry to climate in a forested, nitrogen-saturated catchment recovering from acidification. Ecol. Indic. 2016, 63, 196–208. [Google Scholar] [CrossRef]
- Kaňa, J.; Kopáček, J.; Tahovská, K.; Šantrůčková, H. Tree dieback and related changes in nitrogen dynamics modify the concentrations and proportions of cations on soil sorption complex. Ecol. Indic. 2019, 97, 319–328. [Google Scholar] [CrossRef]
- Matějka, K. Disturbance-induced changes in the plant biomass in forests near Plešné and Čertovo Lakes. J. For. Sci. 2015, 61, 156–168. [Google Scholar] [CrossRef]
- Tahovská, K.; Čapek, P.; Šantrůčková, H.; Kaňa, J.; Kopáček, J. Measurement of in situ Phosphorus Availability in Acidified Soils using Iron-Infused Resin. Commun. Soil Sci. Plant Anal. 2016, 47, 487–494. [Google Scholar] [CrossRef]
- Kopáček, J.; Borovec, J.; Hejzlar, J.; Porcal, P. Spectrophotometric determination of iron, aluminum, and phosphorus in soil and sediment extracts after their nitric and perchloric acid digestion. Commun. Soil Sci. Plant Anal. 2001, 32, 1431–1443. [Google Scholar] [CrossRef]
- Cappo, K.A.; Blume, L.J.; Raab, G.A.; Bartz, J.K.; Engels, J.L. Analytical Methods Manual for the Direct/Delayed Response Project Soil Survey; US EPA: Las Vegas, NV, USA, 1987. [Google Scholar]
- Kopáček, J.; Kaňa, J.; Šantrůčková, H.; Picek, T.; Stuchlík, E. Chemical and Biochemical Characteristics of Alpine Soils in the Tatra Mountains and their Correlation with Lake Water Quality. Water Air Soil Pollut. 2004, 153, 307–328. [Google Scholar] [CrossRef]
- Wolf, A.M.; Baker, D.E. Colorimetric method for phosphorus measurement in ammonium oxalate soil extracts. Commun. Soil Sci. Plant Anal. 1990, 21, 2257–2263. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. Microbial biomass measurements in forest soils—Determination of Kc values and tests of hypotheses to explain the failure of the chloroform fumigation incubation method in acid soils. Soil Biol. Biochem. 1987, 19, 689–696. [Google Scholar] [CrossRef]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform fumigation and the release of soil-nitrogen—A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Brookes, P.C.; Powlson, D.S.; Jenkinson, D.S. Measurement of microbial biomass phosphorus in soil. Soil Biol. Biochem. 1982, 14, 319–329. [Google Scholar] [CrossRef]
- Bárta, J.; Šlajsová, P.; Tahovská, K.; Picek, T.; Šantrůčková, H. Different temperature sensitivity and kinetics of soil enzymes indicate seasonal shifts in C, N and P nutrient stoichiometry in acid forest soil. Biogeochemistry 2014, 117, 525–537. [Google Scholar] [CrossRef]
- Tahovská, K.; Choma, M.; Kaštovská, E.; Oulehle, F.; Bárta, J.; Šantrůčková, H.; Moldan, F. Positive response of soil microbes to long-term nitrogen input in spruce forest: Results from Gårdsjön whole-catchment N-addition experiment. Soil Biol. Biochem. 2020, 143, 107732. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Choma, M.; Tahovská, K.; Kaštovská, E.; Bárta, J.; Růžek, M.; Oulehle, F. Bacteria but not fungi respond to soil acidification rapidly and consistently in both a spruce and beech forest. Fems Microbiol. Ecol. 2020, 96, fiaa174. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Edgar, R. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2024, 52, D791–D797. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- R Core Team. R: A Lanaguage and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Michael, K.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.B.; Simpson, G.; Solymos, P.; et al. Vegan Community Ecology Package Version 2.5-7 November 2020. 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 5 August 2024).
- Lenth, R.V. Emmeans: Estimated Marginal Means, aka Least-Squares Means. 2022. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Choma, M.; Šamonil, P.; Kaštovská, E.; Bárta, J.; Tahovská, K.; Valtera, M.; Šantrůčková, H. Soil Microbiome Composition along the Natural Norway Spruce Forest Life Cycle. Forests 2021, 12, 410. [Google Scholar] [CrossRef]
- Nierman, W.C.; Feldblyum, T.V.; Laub, M.T.; Paulsen, I.T.; Nelson, K.E.; Eisen, J.; Heidelberg, J.F.; Alley, M.R.K.; Ohta, N.; Maddock, J.R.; et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2001, 98, 4136–4141. [Google Scholar] [CrossRef]
- Nixon, S.L.; Daly, R.A.; Borton, M.A.; Solden, L.M.; Welch, S.A.; Cole, D.R.; Mouser, P.J. Genome-Resolved Metagenomics Extends the Environmental Distribution of the Verrucomicrobia Phylum to the Deep Terrestrial Subsurface. Msphere 2019, 4, 10-1128. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Suzina, N.E.; Liesack, W.; Dedysh, S.N. Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemo-organotroph from subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 2010, 60, 301–306. [Google Scholar] [CrossRef]
- Petters, S.; Söllinger, A.; Bengtsson, M.M.; Urich, T. The soil microbial food web revisited with metatranscriptomics—Predatory Myxobacteria as keystone taxon? bioRxiv 2018, 373365. [Google Scholar] [CrossRef]
- Yu, F.; Liang, J.-F.; Song, J.; Wang, S.-K.; Lu, J.-K. Bacterial Community Selection of Russula griseocarnosa Mycosphere Soil. Front. Microbiol. 2020, 11, 347. [Google Scholar] [CrossRef]
- Peeters, C.; De Canck, E.; Cnockaert, M.; De Brandt, E.; Snauwaert, C.; Verheyde, B.; Depoorter, E.; Spilker, T.; LiPuma, J.J.; Vandamme, P. Comparative Genomics of Pandoraea, a Genus Enriched in Xenobiotic Biodegradation and Metabolism. Front. Microbiol. 2019, 10, 2556. [Google Scholar] [CrossRef]
- Ofek, M.; Hadar, Y.; Minz, D. Ecology of Root Colonizing Massilia (Oxalobacteraceae). PLoS ONE 2012, 7, e40117. [Google Scholar] [CrossRef]
- Uroz, S.; Oger, P.; Morin, E.; Frey-Klett, P. Distinct Ectomycorrhizospheres Share Similar Bacterial Communities as Revealed by Pyrosequencing-Based Analysis of 16S rRNA Genes. Appl. Environ. Microbiol. 2012, 78, 3020–3024. [Google Scholar] [CrossRef]
- Talbot, J.M.; Bruns, T.D.; Smith, D.P.; Branco, S.; Glassman, S.I.; Erlandson, S.; Vilgalys, R.; Peay, K.G. Independent roles of ectomycorrhizal and saprotrophic communities in soil organic matter decomposition. Soil Biol. Biochem. 2013, 57, 282–291. [Google Scholar] [CrossRef]
- Kleber, M.; Lehmann, J. Humic Substances Extracted by Alkali Are Invalid Proxies for the Dynamics and Functions of Organic Matter in Terrestrial and Aquatic Ecosystems. J. Environ. Qual. 2019, 48, 207–216. [Google Scholar] [CrossRef]
- Firsching, B.-M.; Claassen, N. Root phosphatase activity and soil organic phosphorus utilization by Norway spruce [Picea abies (L.) Karst.]. Soil Biol. Biochem. 1996, 28, 1417–1424. [Google Scholar] [CrossRef]
- Mooshammer, M.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014, 5, 22. [Google Scholar] [CrossRef]
- Zhang, J.; Elser, J.J. Carbon:Nitrogen:Phosphorus Stoichiometry in Fungi: A Meta-Analysis. Front Microbiol 2017, 8, 1281. [Google Scholar] [CrossRef]
- Mouginot, C.; Kawamura, R.; Matulich, K.L.; Berlemont, R.; Allison, S.D.; Amend, A.S.; Martiny, A.C. Elemental stoichiometry of Fungi and Bacteria strains from grassland leaf litter. Soil Biol. Biochem. 2014, 76, 278–285. [Google Scholar] [CrossRef]
- Kornberg, A. Inorganic polyphosphate: Toward making a forgotten polymer unforgettable. J. Bacteriol. 1995, 177, 491–496. [Google Scholar] [CrossRef]
- Čapek, P.; Tupá, A.; Choma, M. Exploring polyphosphates in soil: Presence, extractability, and contribution to microbial biomass phosphorus. Biol. Fertil. Soils 2024, 60, 667–680. [Google Scholar] [CrossRef]
- Gerlitz, T.G.M. Effects of aluminium on polyphosphate mobilization of the ectomycorrhizal fungus Suillus bovinus. Plant Soil 1996, 178, 133–140. [Google Scholar] [CrossRef]
- Kopáček, J.; Bače, R.; Choma, M.; Hejzlar, J.; Kaňa, J.; Oulehle, F.; Porcal, P.; Svoboda, M.; Tahovská, K. Carbon and nutrient pools and fluxes in unmanaged mountain Norway spruce forests, and losses after natural tree dieback. Sci. Total Environ. 2023, 903, 166233. [Google Scholar] [CrossRef] [PubMed]
- Choma, M.; Bače, R.; Čapek, P.; Kaňa, J.; Kaštovská, E.; Tahovská, K.; Kopáček, J. Surviving trees are key elements in the fate of ectomycorrhizal community after severe bark-beetle forest disturbance. FEMS Microbiol. Ecol. 2023, 99, fiad082. [Google Scholar] [CrossRef]
- Kopáček, J.; Bače, R.; Hejzlar, J.; Kaňa, J.; Kučera, T.; Matějka, K.; Porcal, P.; Turek, J. Changes in microclimate and hydrology in an unmanaged mountain forest catchment after insect-induced tree dieback. Sc. Total Environ. 2020, 720, 137518. [Google Scholar] [CrossRef]



| PL | CT | Initial Site Difference | Incubation Effect | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Litter | Topsoil | Litter | Topsoil | |||||||||
| Initial | Final | Initial | Final | Initial | Final | Initial | Final | F | p | F | p< | |
| CMB µmol g−1 | 841 (116) | 354 (65) * | 223 (31) | 151 (2.7) * | 696 (65) | 331 (28) * | 319 (138) | 127 (14) * | ns | 172 | 0.001 | |
| NMB µmol g−1 | 68 (6.0) | 30 (6.2) * | 16 (2.0) | 14 (0.69) | 51 (2.4) | 28 (3.6) * | 24 (12) | 10 (2.1) * | 90 | 0.001 | ||
| PMB µmol g−1 | 5.4 (0.23) | 12 (3.4) * | 2.0 (0.22) | 8.1 (1.7) * | 4.6 (0.19) | 10 (1.3) * | 2.4 (0.20) | 7.3 (1.2) * | 395 | 0.001 | ||
| CMB/NMB ratio | 12 (1.2) | 12 (0.7) | 14 (0.6) | 10 (0.5) | 14 (1.5) | 12 (0.7) | 14 (1.2) | 13 (1.4) | ns | 24 | 0.001 | |
| NMB/PMB ratio | 12 (1.2) | 3 (0.6) | 8 (0.4) | 2 (0.4) | 11 (0.9) | 3 (0.7) | 10 (4.2) | 1 (0.3) | ns | 613 | 0.001 | |
| specific ENZ activity nmol substrate (µmol CMB)−1 h−1 | 3.0 (0.27) | 17 (4.8) | 6.6 (2.3) | 16 (3.6) | 6.1 (1.1) | 14 (1.6) | 4.4 (0.9) | 18 (2.7) | ns | 185 | 0.001 | |
| fungal 18S rDNA 108 copies g−1 | 6.4 (2.0) | 6.5 (4.9) | 1.1 (0.16) | 1.6 (0.68) | 4.3 (1.8) | 8.1 (2.1) | 1.3 (0.45) | 2.0 (0.45) | ns | 7.5 | 0.01 | |
| bacterial 16S rDNA 109 copies g−1 | 6.7 (2.5) | 3.9 (0.67) | 6.1 (1.0) | 5.8 (1.2) | 4.7 (0.57) | 5.9 (1.0) | 4.7 (0.91) | 4.4 (1.1) | 8.9 | 0.01 | ns | |
| bacteria/fungi ratio | 12 | 8.5 | 55 | 39 | 12 | 7.5 | 39 | 23 | ns | 6.4 | 0.02 | |
| TOC µmol g−1 | 88 (4.9) c | 38 (6.2) * | 39 (2.5) a | 27 (1.8) * | 65 (3.6) b | 42 (4.1) * | 43 (8.1) b | 33 (3.2) | 26 | <0.001 | 182 | 0.001 |
| N-NH4 (H2O) nmol g−1 | 481 (105) b | 910 (771) | 147 (23) a | 1523 (965) * | 579 (185) b | 1435 (444) | 191 (23) a | 3066 (1611) * | 4.9 | 0.042 | 100 | 0.001 |
| N-NO3 (H2O) nmol g−1 | 213 (47) a | 48 (19) * | 170 (48) a | 58 (29) * | 300 (16) | 13 (3.3) * | 328 (8.4) b | 26 (9.0) * | 46 | <0.001 | 501 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahovská, K.; Choma, M.; Čapek, P.; Kaštovská, E.; Kaňa, J.; Kopáček, J. Increased Saprotrophic Activity and Phosphate Leaching Following Forest Soil Decomposition without Root Access. Forests 2024, 15, 1378. https://doi.org/10.3390/f15081378
Tahovská K, Choma M, Čapek P, Kaštovská E, Kaňa J, Kopáček J. Increased Saprotrophic Activity and Phosphate Leaching Following Forest Soil Decomposition without Root Access. Forests. 2024; 15(8):1378. https://doi.org/10.3390/f15081378
Chicago/Turabian StyleTahovská, Karolina, Michal Choma, Petr Čapek, Eva Kaštovská, Jiří Kaňa, and Jiří Kopáček. 2024. "Increased Saprotrophic Activity and Phosphate Leaching Following Forest Soil Decomposition without Root Access" Forests 15, no. 8: 1378. https://doi.org/10.3390/f15081378
APA StyleTahovská, K., Choma, M., Čapek, P., Kaštovská, E., Kaňa, J., & Kopáček, J. (2024). Increased Saprotrophic Activity and Phosphate Leaching Following Forest Soil Decomposition without Root Access. Forests, 15(8), 1378. https://doi.org/10.3390/f15081378







