Homogeneous Selection Mediated by Nitrate Nitrogen Regulates Fungal Dynamics in Subalpine Forest Soils Subjected to Simulated Restoration
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Natural Soil Sampling
2.2. Experimental Procedures and Cultivated Soil Sampling
2.3. Soil Property Measurement
2.4. DNA Extraction, Amplification, and Sequencing
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
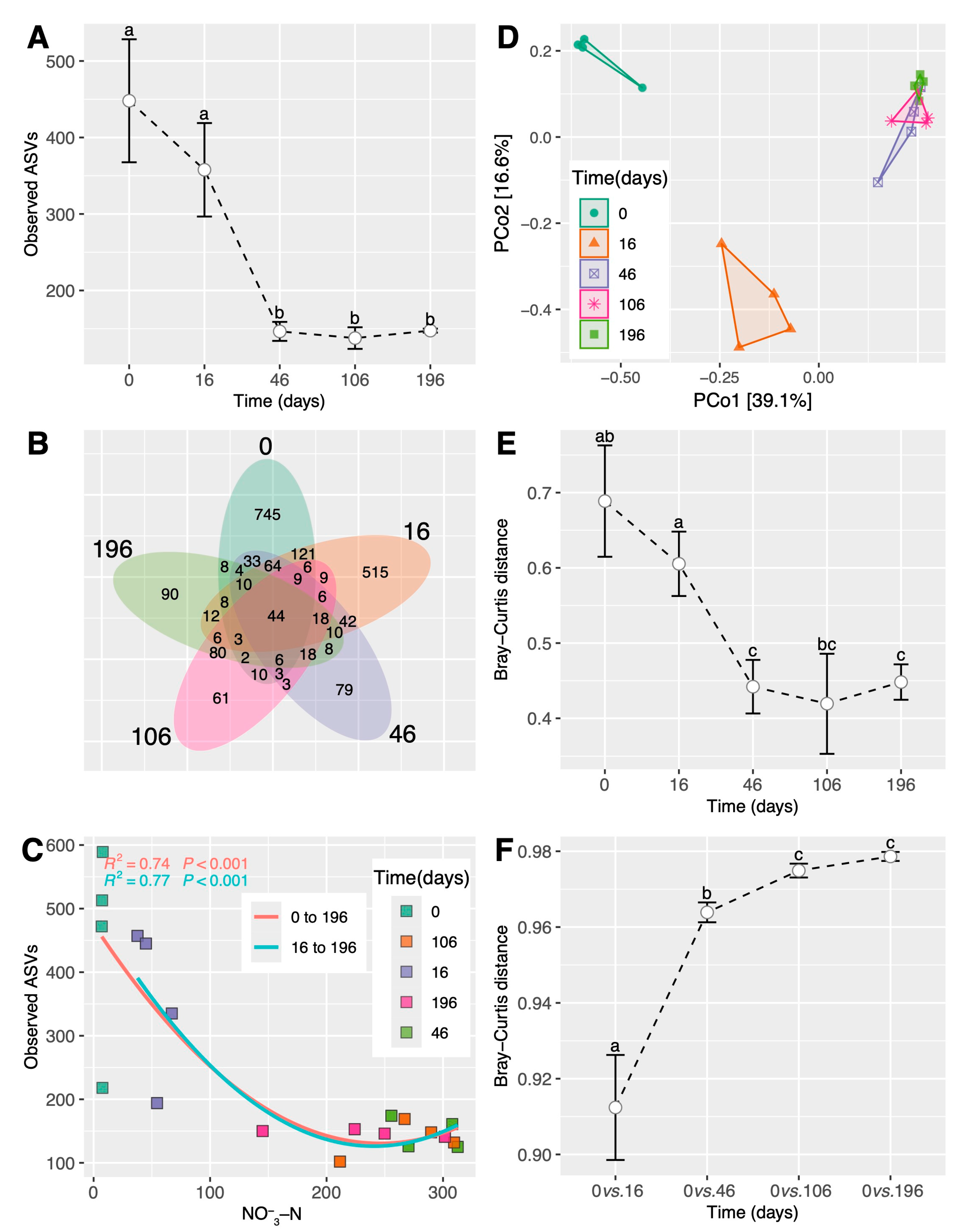
3.1. Shifts in Soil Conditions during the Simulated ESSS-Based Restoration
3.2. Shifts in Fungal Diversity and Community during the Simulated ESSS-Based Restoration
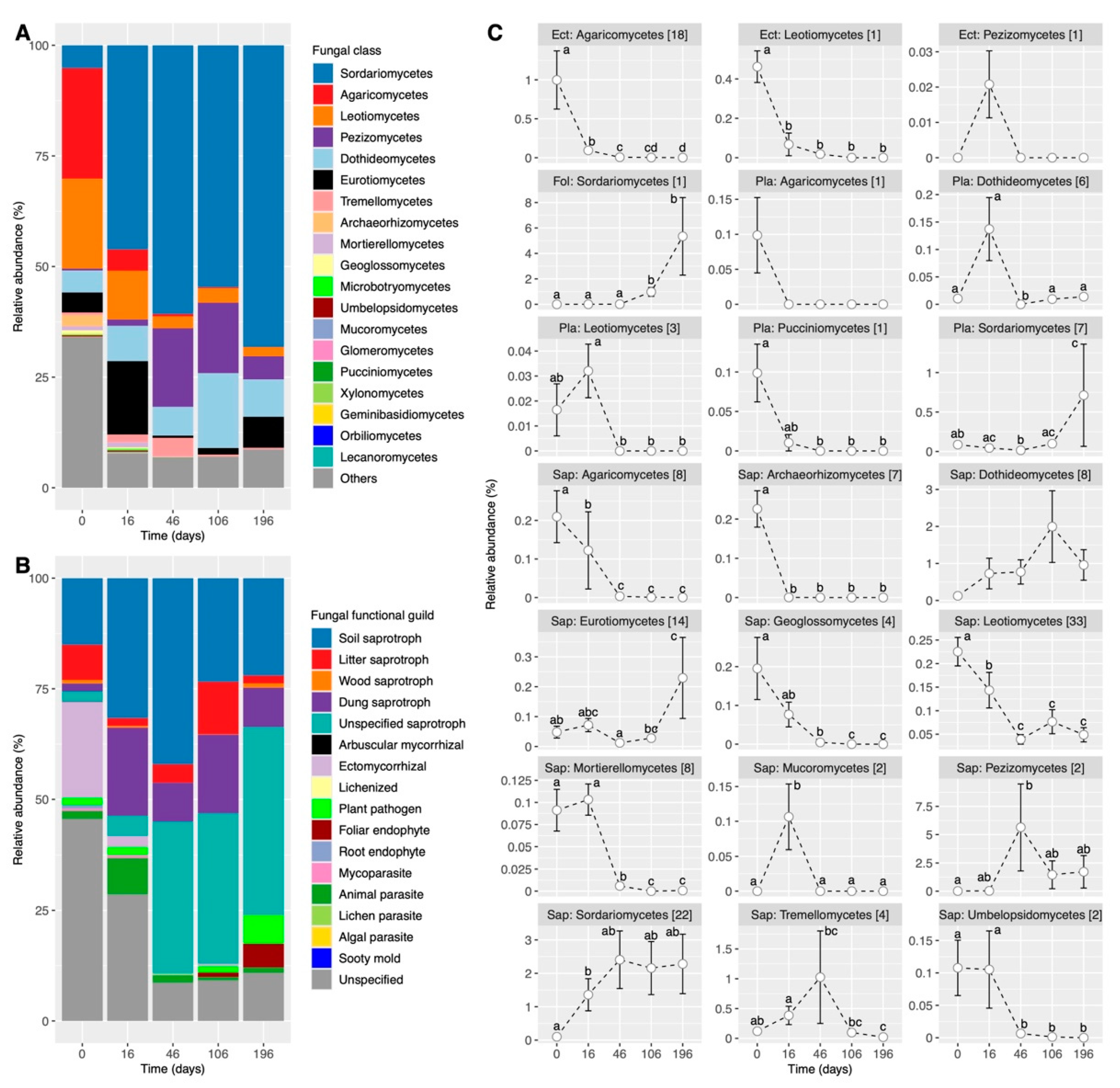
3.3. Shifts in the Composition of Functional Guilds during the Simulated ESSS-Based Restoration
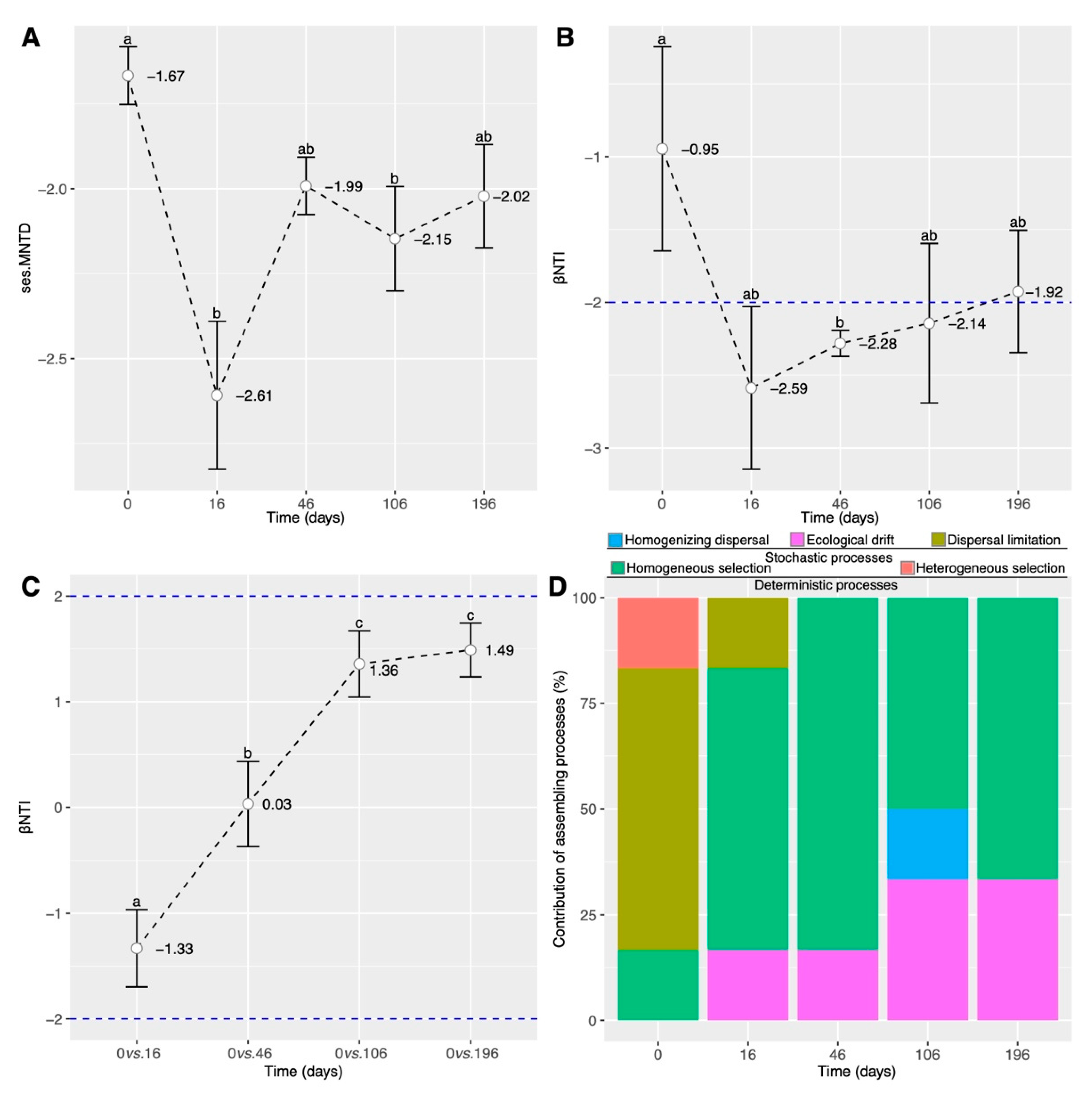
3.4. Shifts in Fungal Community Assembly Processes during the Simulated ESSS-Based Restoration
4. Discussion
4.1. Fungal Dynamics Are Mainly Characterized by Ectomycorrhiza, Pathogens, and Saprotrophs
4.2. Ectomycorrhiza Are Depleted, but Saprotrophs and Pathogens Are Enriched over Time
4.3. Homogeneous Selection Driven by Nitrate Nitrogen Content Regulates Soil Fungal Dynamics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, C.; Liao, H.; Li, D.; Jing, Y. The fungal functional guilds at the early-stage restoration of subalpine forest soils disrupted by highway construction in Southwest China. Forests 2024, 15, 636. [Google Scholar] [CrossRef]
- Meyer, M.D. Natural Range of Variation of Subalpine Forests in the Bioregional Assessment Area; US Forest Service, Pacific Southwest Region: Washington, DC, USA, 2013. [Google Scholar]
- Banerjee, P.; Ghose, M.K.; Pradhan, R. Analytic hierarchy process based spatial biodiversity impact assessment model of highway broadening in Sikkim Himalaya. Geocarto Int. 2020, 35, 470–493. [Google Scholar] [CrossRef]
- Van Der Ree, R.; Smith, D.J.; Grilo, C. The ecological effects of linear infrastructure and traffic: Challenges and opportunities of rapid global growth. In Handbook of Road Ecology; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar]
- Xu, H.; Li, T.B.; Chen, J.N.; Liu, C.N.; Zhou, X.h.; Xia, L. Characteristics and applications of ecological soil substrate for rocky slope vegetation in cold and high-altitude areas. Sci. Total Environ. 2017, 609, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Ai, S.; Chen, J.; Gao, D.; Ai, Y. Distribution patterns and drivers of artificial soil bacterial community on cut-slopes in alpine mountain area of southwest China. Catena 2020, 194, 104695. [Google Scholar] [CrossRef]
- Fu, D.; Yang, H.; Wang, L.; Yang, S.; Li, R.; Zhang, W.; Ai, X.; Ai, Y. Vegetation and soil nutrient restoration of cut slopes using outside soil spray seeding in the plateau region of southwestern China. J. Environ. Manag. 2018, 228, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Ramaji, A.E. A review on the soil stabilization using low-cost methods. J. Appl. Sci. Res. 2012, 8, 2193–2196. [Google Scholar]
- Liao, H.; Li, C.; Ai, S.; Li, X.; Ai, X.; Ai, Y. A simulated ecological restoration of bare cut slope reveals the dosage and temporal effects of cement on ecosystem multifunctionality in a mountain ecosystem. J. Environ. Manag. 2023, 325, e116672. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Sheng, M.; Liu, J.; Ai, X.; Li, C.; Ai, S.; Ai, Y. Soil N availability drives the shifts of enzyme activity and microbial phosphorus limitation in the artificial soil on cut slope in southwestern China. Environ. Sci. Pollut. Res. 2021, 28, 33307–33319. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, J.; Meng, Z.; Xu, R.; Chen, J.; Zhang, Y.; Hu, T. Fertility-related interplay between fungal guilds underlies plant richness–productivity relationships in natural grasslands. New Phytol. 2020, 226, 1129–1143. [Google Scholar] [CrossRef]
- Li, J.; Delgado Baquerizo, M.; Wang, J.T.; Hu, H.W.; Cai, Z.J.; Zhu, Y.N.; Singh, B.K. Fungal richness contributes to multifunctionality in boreal forest soil. Soil Biol. Biochem. 2019, 136, 107526. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Ngugi, M.R.; Fechner, N.; Neldner, V.J.; Dennis, P.G. Successional dynamics of soil fungal diversity along a restoration chronosequence post-coal mining. Restor. Ecol. 2020, 28, 543–552. [Google Scholar] [CrossRef]
- Wang, K.; Bi, Y.; Cao, Y.; Peng, S.; Christie, P.; Ma, S.; Zhang, J.; Xie, L. Shifts in composition and function of soil fungal communities and edaphic properties during the reclamation chronosequence of an open-cast coal mining dump. Sci. Total Environ. 2021, 767, 144465. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jia, Z.; Yuan, Y.; Cheng, X.; Shi, J.; Tang, X.; Wang, Y.; Peng, X.; Dong, Y.; Ma, S.; et al. Effects of mineral-solubilizing microbial strains on the mechanical responses of roots and root-reinforced soil in external-soil spray seeding substrate. Sci. Total Environ. 2020, 723, 138079. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, J.; Ai, X.; Li, R.; Ai, Y.; Li, W. The texture, structure and nutrient availability of artificial soil on cut slopes restored with OSSS–Influence of restoration time. J. Environ. Manag. 2017, 200, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Li, Z.; Han, R. Statistical evaluation of sprayed synthetic soils amended with four additive factors used in high- and cut rock slopes. Environ. Eng. Sci. 2016, 34, 281–290. [Google Scholar] [CrossRef]
- Zhao, X.; Li, Z.; Robeson, M.D.; Hu, J.; Zhu, Q. Application of erosion-resistant fibers in the recovery of vegetation on steep slopes in the Loess Plateau of China. Catena 2018, 160, 233–241. [Google Scholar] [CrossRef]
- Liao, H.; Li, C.; Ai, Y.; Li, X. Soil bacterial responses to disturbance are enlarged by altitude in a mountain ecosystem. J. Soils Sediments 2023, 23, 3820–3831. [Google Scholar] [CrossRef]
- Li, C.; Liao, H.; Li, D.; Jing, Y. Abundant species govern the altitude patterns of bacterial community in natural and disturbed subalpine forest soils. Diversity 2024, 16, 242. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhao, Q.; Lai, Q.; Xu, G.; Chen, X. Cognition and practice on technological innovation of ecological restoration of engineering slope soil in South China. Soils 2017, 49, 643–650. [Google Scholar]
- Jin, X.; Liu, Y.; Hu, W.; Wang, G.; Kong, Z.; Wu, L.; Ge, G. Soil bacterial and fungal communities and the associated nutrient cycling responses to forest conversion after selective logging in a subtropical forest of China. For. Ecol. Manag. 2019, 444, 308–317. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Van Der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.G.A.; Martin, F.M.; Selosse, M.A.; Sanders, I.R. Mycorrhizal ecology and evolution: The past, the present, and the future. New Phytol. 2015, 205, 1406–1423. [Google Scholar] [CrossRef] [PubMed]
- García-Guzmán, G.; Heil, M. Life histories of hosts and pathogens predict patterns in tropical fungal plant diseases. New Phytol. 2013, 201, 1106–1120. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Mitschunas, N. Fungal effects on seed bank persistence and potential applications in weed biocontrol: A review. Basic Appl. Ecol. 2008, 9, 191–203. [Google Scholar] [CrossRef]
- Mahedi, M.; Cetin, B.; White, D.J. Cement, lime, and fly ashes in stabilizing expansive soils: Performance evaluation and comparison. J. Mater. Civ. Eng. 2020, 32, 04020177. [Google Scholar] [CrossRef]
- Větrovský, T.; Kohout, P.; Kopecký, M.; Machac, A.; Man, M.; Bahnmann, B.D.; Brabcová, V.; Choi, J.; Meszárošová, L.; Human, Z.R.; et al. A meta-analysis of global fungal distribution reveals climate-driven patterns. Nat. Commun. 2019, 10, 5142. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, C.; Tu, B.; Kou, Y.; Li, X. Species pool and local ecological assembly processes shape the β-diversity of diazotrophs in grassland soils. Soil Biol. Biochem. 2021, 160, 108338. [Google Scholar] [CrossRef]
- Meiying, X.; Qin, Z.; Chunyu, X.; Yuming, Z.; Sun, G.; Jun, G.; Yuan, T.; Jizhong, Z.; Zhili, H. Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. ISME J. 2014, 8, 1932–1944. [Google Scholar]
- Nygren, C.M.R.; Eberhardt, U.; Karlsson, M.; Parrent, J.L.; Lindahl, B.D.; Taylor, A.F.S. Growth on nitrate and occurrence of nitrate reductase-encoding genes in a phylogenetically diverse range of ectomycorrhizal fungi. New Phytol. 2008, 180, 875–889. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, Y.; Zhao, W.; He, H.; Li, D.; He, W.; Liu, Q.; Yin, H. Seasonal variations in the soil amino acid pool and flux following the conversion of a natural forest to a pine plantation on the eastern Tibetan Plateau, China. Soil Biol. Biochem. 2017, 105, 1–11. [Google Scholar] [CrossRef]
- GB 175–2007; Common Portland Cement. Standards Press of China: Beijing, China, 2007.
- NB/T 35082-2016; Technical Code for Eco-restoration of Vegetation Concrete On Steep Slope of Hydropower Projects. China Electric Power Press: Beijing, China, 2016.
- Chunjuan, G.; Lihua, J.; Guangning, W.; Jing, l.; Qiuyue, H.; Guizhai, Z.; Huiqun, R.; Cong, W. Effects of different fungicides on seed germination of tall fescue in slope plants and soil properties. J. Shandong Agric. Eng. Univ. 2024, 41. [Google Scholar] [CrossRef]
- He, H.; Yu, L.; Yang, X.; Luo, L.; Liu, J.; Chen, J.; Kou, Y.; Zhao, W.; Liu, Q. Effects of different soils on the biomass and photosynthesis of rumex nepalensis in Subalpine Region of Southwestern China. Forests 2022, 13, 73. [Google Scholar] [CrossRef]
- Bao, S. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter; Soil Science Society of America, American Society of Agronomy: Washington, DC, USA, 1996. [Google Scholar]
- Bremner, J.M. Total Nitrogen. In Methods of Soil Analysis: Part 2. Chemical and Microbiological Properties; Black, C.A., Evans, D.D., Dinauer, R.C., Eds.; American Society of Agronomy, Soil Science Society of America: Washington, DC, USA, 1965. [Google Scholar]
- Wang, Y.; Li, C.; Kou, Y.; Wang, J.; Tu, B.; Li, H.; Li, X.; Wang, C.; Yao, M. Soil pH is a major driver of soil diazotrophic community assembly in Qinghai-Tibet alpine meadows. Soil Biol. Biochem. 2017, 115, 547–555. [Google Scholar] [CrossRef]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; U.S. Department of Agriculture: Washington, DC, USA, 1954. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Callahan, B.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth's multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.; Schigel, D.; Kennedy, P.; Picard, K.T.; Glöckner, F.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Li, C.; Liu, C.; Li, H.; Liao, H.; Xu, L.; Yao, M.; Li, X. The microgeo: An R package rapidly displays the biogeography of soil microbial community traits on maps. FEMS Microbiol. Ecol. 2024, 100, fiae087. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.L.; Yuan, M.T.; Wu, L.W.; Zhang, Y.; Guo, X.; Zhou, X.S.; Yang, Y.F.; Arkin, A.P.; Firestone, M.K.; Zhou, J.Z. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, e4717. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Online Library, Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Rodriguez Ramos, J.C.; Cale, J.A.; Cahill, J.F.; Simard, S.W.; Karst, J.; Erbilgin, N. Changes in soil fungal community composition depend on functional group and forest disturbance type. New Phytol. 2020, 229, 1105–1117. [Google Scholar] [CrossRef]
- Rähn, E.; Tedersoo, L.; Adamson, K.; Drenkhan, T.; Sibul, I.; Lutter, R.; Anslan, S.; Pritsch, K.; Drenkhan, R. Rapid shift of soil fungal community compositions after clear-cutting in hemiboreal coniferous forests. For. Ecol. Manag. 2023, 544, 121211. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Fernández, N.; Knoblochová, T.; Kohout, P.; Janoušková, M.; Cajthaml, T.; Frouz, J.; Rydlová, J. Asymmetric interaction between two mycorrhizal fungal guilds and consequences for the establishment of their host plants. Front. Plant Sci. 2022, 13, 873204. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M. Mycorrhizal types differ in ecophysiology and alter plant nutrition and soil processes. Biol. Rev. 2019, 94, 1857–1880. [Google Scholar] [CrossRef] [PubMed]
- Peay, K.G. The mutualistic niche: Mycorrhizal symbiosis and community dynamics. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 143–164. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, Z.; Sun, H.; Yang, W.; Xu, H. The response patterns of arbuscular mycorrhizal and ectomycorrhizal symbionts under elevated CO2: A meta-analysis. Front. Microbiol. 2018, 9, 1248. [Google Scholar] [CrossRef]
- Li, S.p.; Cadotte, M.W.; Meiners, S.J.; Hua, Z.s.; Jiang, L.; Shu, W.S. Species colonisation, not competitive exclusion, drives community overdispersion over long-term succession. Ecol. Lett. 2015, 18, 964–973. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Burgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.; et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, H.; Li, D.; Li, C. Homogeneous Selection Mediated by Nitrate Nitrogen Regulates Fungal Dynamics in Subalpine Forest Soils Subjected to Simulated Restoration. Forests 2024, 15, 1385. https://doi.org/10.3390/f15081385
Liao H, Li D, Li C. Homogeneous Selection Mediated by Nitrate Nitrogen Regulates Fungal Dynamics in Subalpine Forest Soils Subjected to Simulated Restoration. Forests. 2024; 15(8):1385. https://doi.org/10.3390/f15081385
Chicago/Turabian StyleLiao, Haijun, Dehui Li, and Chaonan Li. 2024. "Homogeneous Selection Mediated by Nitrate Nitrogen Regulates Fungal Dynamics in Subalpine Forest Soils Subjected to Simulated Restoration" Forests 15, no. 8: 1385. https://doi.org/10.3390/f15081385
APA StyleLiao, H., Li, D., & Li, C. (2024). Homogeneous Selection Mediated by Nitrate Nitrogen Regulates Fungal Dynamics in Subalpine Forest Soils Subjected to Simulated Restoration. Forests, 15(8), 1385. https://doi.org/10.3390/f15081385



