Timber Tracking of Jacaranda copaia from the Amazon Forest Using DNA Fingerprinting
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Extraction and SNP Analysis
2.3. Genetic Diversity Analysis
2.4. Bayesian Clustering Analysis
2.5. Genetic Differentiation among Populations, Countries, and Crusters
2.6. Genetic Assignment Analysis
3. Results
3.1. Genetic Diversity
3.2. Bayesian Cluster
3.3. Population Differentiation
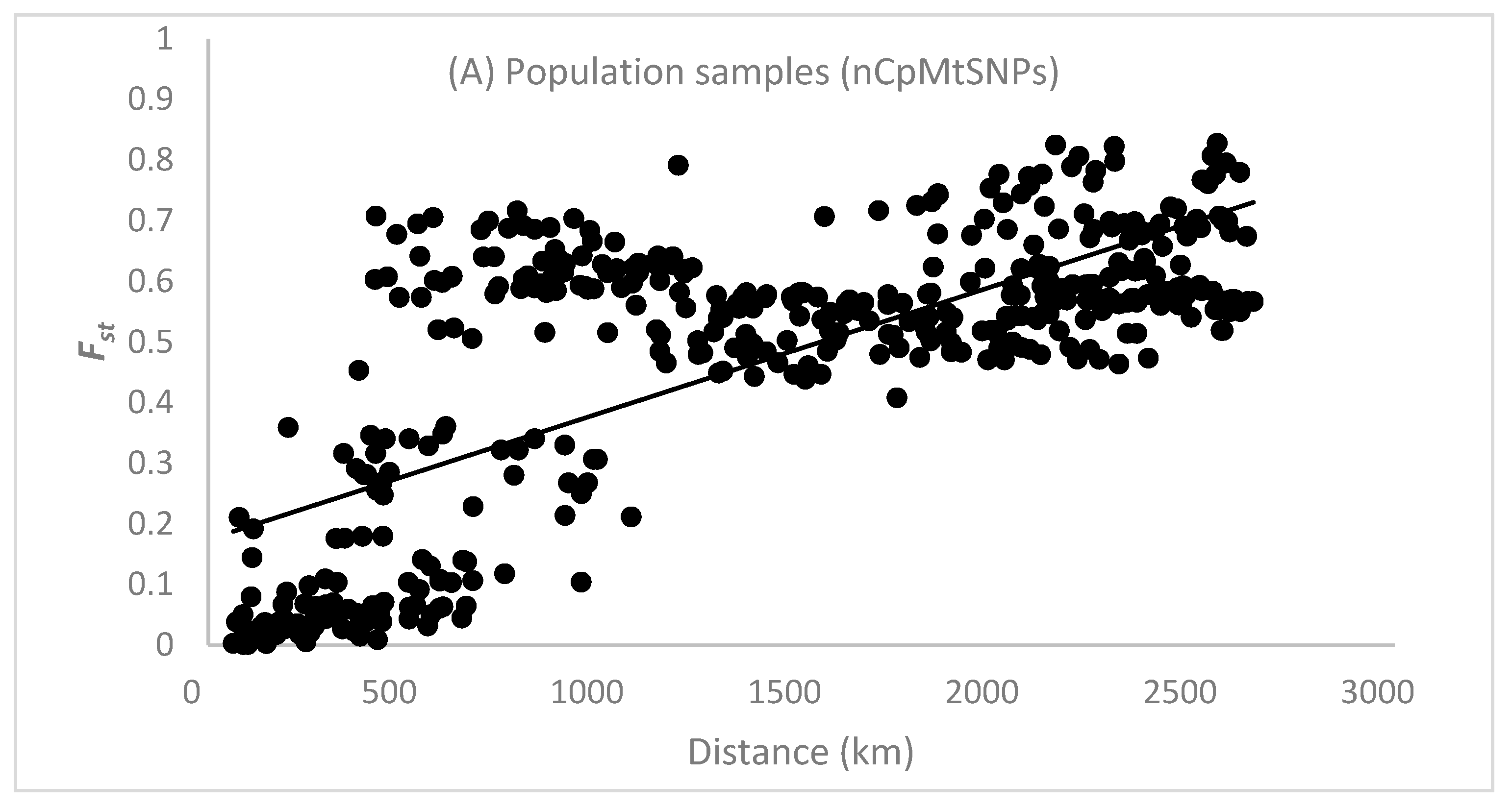
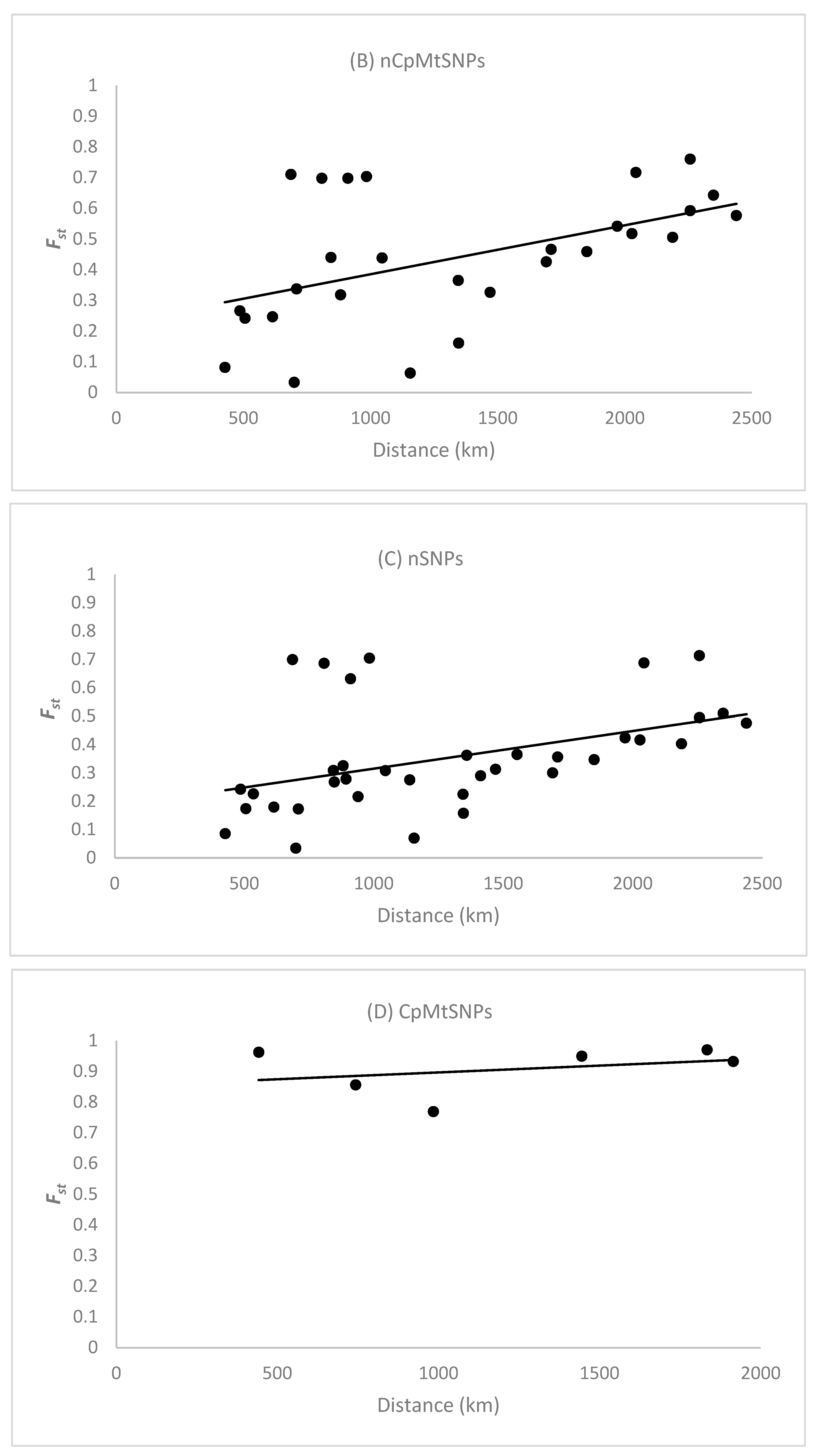
3.4. Isolation by Distance
3.5. Genetic Assignment
4. Discussion
4.1. Genetic Diversity
4.2. Population Genetic Differentiation
4.3. Genetic Assignment and Practical Applications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Degen, D.; Ward, S.E.; Lemes, M.R.; Navarro, C.; Cavers, S.; Sebbenn, A.M. Verifying the geographic origin of mahogany (Swietenia macrophylla King) with DNA-fingerprints. Forensic Sci. Int. Genet. 2013, 7, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.; Lin, C.; Ju, L. Tracking the geographical origin of timber by DNA fingerprinting: A study of the endangered species Cinnamomum kanehirae in Taiwan. Holzforschung 2017, 71, 853–862. [Google Scholar] [CrossRef]
- CITES. Comércio Internacional das Espécies da Flora e Fauna Selvagens em Perigo de Extinção. 2021. Available online: https://cites.org/esp/app/appendices.php (accessed on 1 June 2023).
- Lescuyer, G.; Ndotit, S.; Ndong, L.B.B.; Tsanga, R.; Cerutti, P.O. Policy Options for Improved Integration of Domestic Timber Markets under the Voluntary Partnership Agreement (VPA) Regime in Gabon; Center for International Forestry Research (CIFOR): Bogor, Indonesia, 2014. [Google Scholar]
- Federal Police Brazil. GOV. 2022. Available online: https://www.gov.br/pf/pt-br/search?SearchableText=madeira%20ilegal%20extra%C3%ADda%20da%20Amaz%C3%B4nia (accessed on 17 May 2022).
- Farias, E. Amazônia em Chamas: 90% da Madeira Exportada São Ilegais, Diz Polícia Federal. Amazônia Real, September 13. 2019. Available online: https://amazoniareal.com.br/amazonia-em-chamas-90-da-madeira-exportada-sao-ilegais-diz-policia-federal/67 (accessed on 31 July 2023).
- Perez Coelho, M.; Sanz, J.; Cabezudo, M. Analysis of volatile components of oak wood by solvent extraction and direct thermal desorption-gas chromatography-mass spectrometry. J. Chromat. 1997, 778, 427–434. [Google Scholar] [CrossRef]
- Deklerck, V.; Finch, K.; Gasson, P.; Van den Bulcke, J.; Van Acker, J.; Beeckman, H.; Espinoza, E. Comparison of species classification models of mass spectrometry data: Kernel discriminant analysis vs. random forest; A case study of Afrormosia (Pericopsis elata (Harms) Meeuwen). Rapid Commun. Mass Spectrom. 2017, 31, 1582–1588. [Google Scholar] [CrossRef]
- Kagawa, A.; Leavitt, S.W. Stable carbon isotopes of tree rings as a tool to pinpoint the geographic origin of timber. J. Wood Sci. 2010, 56, 175–183. [Google Scholar] [CrossRef]
- Bergo, M.C.J.; Pastore, T.C.M.; Coradin, V.T.R.; Wiedenhoeft, A.C.; Braga, J.W.B. NIRS identification of Swietenia macrophylla is robust across specimens from 27 countries. IAWA J. 2016, 37, 420–430. [Google Scholar] [CrossRef]
- Gasson, P.; Baas, P.; Wheeler, E. Wood anatomy of Cites-listed tree species. IAWA J. 2011, 32, 155–198. [Google Scholar] [CrossRef]
- Moya, R.; Wiemann, M.C.; Olivares, C. Identification of endangered or threatened Costa Rican tree species by wood anatomy and fluorescence activity. Rev. Biol. Trop. 2013, 61, 1113–1156. [Google Scholar]
- Tnah, L.H.; Lee, S.L.; Ng, K.K.S.; Tani, N.; Bhassu, S.; Othman, R.Y. Geographical traceability of an important tropical timber (Neobalanocarpus heimii) inferred from chloroplast DNA. For. Ecol. Manag. 2009, 258, 1918–1923. [Google Scholar] [CrossRef]
- Jolivet, C.; Degen, D. Use of DNA fingerprints to control the origin of sapelli timber (Entandrophragma cylindricum) at the forest concession level in Cameroon. Forensic Sci. Int. Genet. 2012, 6, 487–493. [Google Scholar] [CrossRef]
- Chaves, C.L.; Degen, B.; Pakull, B.; Mader, M.; Honorio, E.; Ruas, P.; Tysklind, N.; Sebbenn, A.M. Assessing the ability of chloroplast and nuclear DNA gene markers to verify the geographic origin of Jatoba (Hymenaea courbaril L.) timber. J. Hered. 2018, 109, 543–552. [Google Scholar] [CrossRef]
- Coronado, E.N.H.; Blanc-Jolivet, C.; Mader, M.; García-Dávila, C.R.; Gomero, D.A.; del Castillo, D.T.; Llampazo, G.F.; Pizango, G.H.; Sebbenn, A.M.; Meyer-Sand, B.R.V.; et al. SNP markers as a successful molecular tool for assessing species identity and geographic origin of trees in the economically important South American legume Genus. Dipteryx. J. Hered. 2020, 111, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Vlam, M.; de Groot, G.A.; Boom, A.; Copini, P.; Laros, I.; Veldhuijzen, K.; Zakamdi, D.; Zuidema, P.A. Developing forensic tools for an African timber: Regional origin is revealed by genetic characteristics, but not by isotopic signature. Biol. Conserv. 2018, 220, 262–271. [Google Scholar] [CrossRef]
- Abeele, S.V.; Hardy, O.J.; Beeckman, H.; Ilondea, B.A.; Janssens, S.B. Genetic markers for species conservation and timber tracking: Development of microsatellite primers for the Tropical African tree species Prioria balsamifera and Prioria oxyphylla. Forests 2019, 10, 1037. [Google Scholar] [CrossRef]
- Loureiro, A.A.; Silva, M.F.; Alencar, J.C. Essências Madeireiras da Amazonia; INPA: Bergen op Zoom, The Netherlands, 1979; Volume 1. [Google Scholar]
- Sampaio, P.T.B.; Barbosa, A.P.; Fernandes, N.P. Ensaio de espaçamento com caroba—Jacaranda copaia (AUBL.) D. Don. Bignonaceae. Acta Amaz. 1989, 9, 383. [Google Scholar] [CrossRef][Green Version]
- Gentry, A.H. Bignoniaceae—Part II (Tribe Tecomeae). Flora Neotrop. 1992, 25, 1–370. [Google Scholar]
- Maues, M.M.; Oliveira, P.E.A.M.; Kanashiro, M. Pollination biology in Jacaranda copaia (Aubl.) D. Don. (Bignoniaceae) at the “Floresta Nacional do Tapajós”, Central Amazon. Rev. Brasil. Bot. 2008, 31, 517–527. [Google Scholar] [CrossRef][Green Version]
- James, T.; Veges, S.; Aldrich, P.; Hamrick, J.L. Mating systems of three tropical dry Forest tree species. Biotropica 1998, 30, 587–594. [Google Scholar] [CrossRef]
- Vinson, C.C.; Kanashiro, M.; Harris, S.A.; Boshier, D.H. Impacts of selective logging on inbreeding and gene flow in two Amazonian timber species with contrasting ecological and reproductive characteristics. Mol. Ecol. 2015, 24, 38–53. [Google Scholar] [CrossRef]
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pediculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef]
- Sebbenn, A.M.; Blanc-Jolivet, C.; Mader, M.; Meyer-Sand, B.R.V.; Paredes-Villanueva, K.; Coronado, E.N.H.; Garcia-Davila, C.; Tysklind, N.; Troispoux, V.; Delcamp, A.; et al. Nuclear and plastidial SNP and INDEL markers for genetic tracking studies of Jacaranda copaia. Conserv. Genet. Resour. 2019, 11, 341–343. [Google Scholar] [CrossRef]
- Degen, B. Application. GDA-NT 2021—A computer program for population genetic data analysis and assignment. Conserv. Genet. Resour. 2022, 14, 347–350. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 1995, 82, 1420–1425. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDI: A versatile computer program to analyses spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Rannala, B.; Mountain, J.L. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 9197–9201. [Google Scholar] [CrossRef]
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2: A software for genetic assignment and first-generation migrant detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef]
- Efron, B. Estimating the error rate of a prediction rule—Improvement on cross validation. J. Am. Stat. Assoc. 1983, 78, 316–331. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Piry, S.; Luikart, G.; Estoup, A.; Solignac, M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 1999, 153, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Fatima, T.; Srivastava, A.; Hanur, V.S.; Rao, S.M. An effective wood DNA extraction protocol for three economic important timber species of India. Am. J. Plant Sci. 2018, 9, 139–149. [Google Scholar] [CrossRef][Green Version]
- Méndez-Cea, B.; Cobo-Simón, I.; Pérez-González, A.; García-García, I.; Linares, J.C.; Rodríguez, F.J.G. DNA extraction and amplification from Pinaceae dry wood. Silvae Genet. 2019, 68, 55–57. [Google Scholar] [CrossRef]
- Murillo-Sánchez, I.E.; López-Albarrán, P.; Santoyo-Pizano, G.; Martínez-Pacheco, M.M.; Velázquez-Becerra, C. Molecular identification of timber species from sawn timber and roundwood. Conserv. Genet. Resour. 2021, 13, 191–200. [Google Scholar] [CrossRef]
- Tysklind, N.; Blanc-Jolivet, C.; Mader, M.; Meyer-Sand, B.R.V.; Paredes-Villanueva, K.; Coronado, E.N.H.; Garcia-Davila, C.; Sebbenn, A.M.; Caron, K.; Troispoux, V.; et al. Development of nuclear and plastid SNP and INDEL markers for population genetic studies and timber traceability of Carapa species. Conserv. Genet. Resour. 2019, 11, 337–339. [Google Scholar] [CrossRef]
- Meyer-Sand, B.R.V.; Blanc-Jolivet, C.; Mader, M.; Paredes-Villanueva, K.; Tysklind, N.; Sebbenn, A.M.; Guichoux, E.; Degen, D. Development of a set of SNP markers for population genetics studies of Ipe (Handroanthus sp.), a valuable tree genus from Latin America. Conserv. Genet. Resour. 2018, 10, 779–781. [Google Scholar] [CrossRef]
- Tnah, L.H.; Lee, S.L.; Ng, K.K.S.; Faridah, Q.-Z.; Faridah-Hanum, I. Forensic DNA profiling of tropical timber species in Peninsular Malaysia. For. Ecol. Manag. 2010, 259, 1436–1446. [Google Scholar] [CrossRef]
- Ng, K.K.; Lee, S.L.; Tnah, L.H.; Nurul-Farhanah, Z.; Ng, C.H.; Lee, C.T.; Tani, N.; Diway, B.; Lai, P.S.; Khoo, E. Forensic timber identification: A case study of a cites listed species, Gonystylus bancanus (Thymelaeaceae). Forensic Sci. Int. Genet. 2016, 23, 197–209. [Google Scholar] [CrossRef]
- Ng, C.H.; Lee, S.L.; Tnah, L.H.; Ng, K.K.S.; Lee, C.T.; Diway, B.; Khoo, E. Geographic origin and individual assignment of Shorea platyclados (Dipterocarpaceae) for forensic identification. PLoS ONE 2017, 12, e0176158. [Google Scholar] [CrossRef]
- Ng, C.H.; Ng, K.K.S.; Lee, S.L.; Zakari, N.-F.; Lee, C.T.; Tnah, L.H. DNA databases of an important tropical timber tree species Shorea leprosula (Dipterocarpaceae) for forensic timber identification. Sci. Rep. 2022, 12, 9546. [Google Scholar] [CrossRef]
- Wahlund, S. Composition of populations and correlation appearances viewed in relation to the studies of inheritance. Hereditas 1928, 11, 65–106. [Google Scholar] [CrossRef]
- Scotti-Saintagne, C.; Dick, C.W.; Caron, H.; Vendramin, G.G.; Troispoux, V.; Sire, P.; Casalis, M.; Buonamici, A.; Valencia, R.; Lemes, M.R.; et al. Amazon diversification and cross-Andean dispersal of the widespread Neotropical tree species Jacaranda copaia (Bignoniaceae). J. Biogeogr. 2013, 40, 707–719. [Google Scholar] [CrossRef]
- Roman, G.; Gangitano, D.; Figueroa, A.; Solano, J.; Anabalón, L.; Houston, R. Use of Eucalyptus DNA profiling in a case of illegal logging. Sci. Justice 2020, 60, 487–494. [Google Scholar] [CrossRef]
- Huang, C.; Chu, F.H.; Huang, Y.-S.; Hung, Y.M.; Tseng, Y.-S.; Pu, C.-E.; Chao, C.-H.; Chou, S.-H.; Liu, S.-H.; You, Y.T.Y.; et al. Development and technical application of SSR-based individual identifcation system for Chamaecyparis taiwanensis against illegal logging convictions. Sci. Rep. 2020, 10, 22095. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Ding, X.; Feng, T.Y.; Chen, T.; Ye, J. Genetic diversity and population structure of Bursaphelenchus xylophilus in central China based on SNP Markers. Forests 2023, 14, 1443. [Google Scholar] [CrossRef]

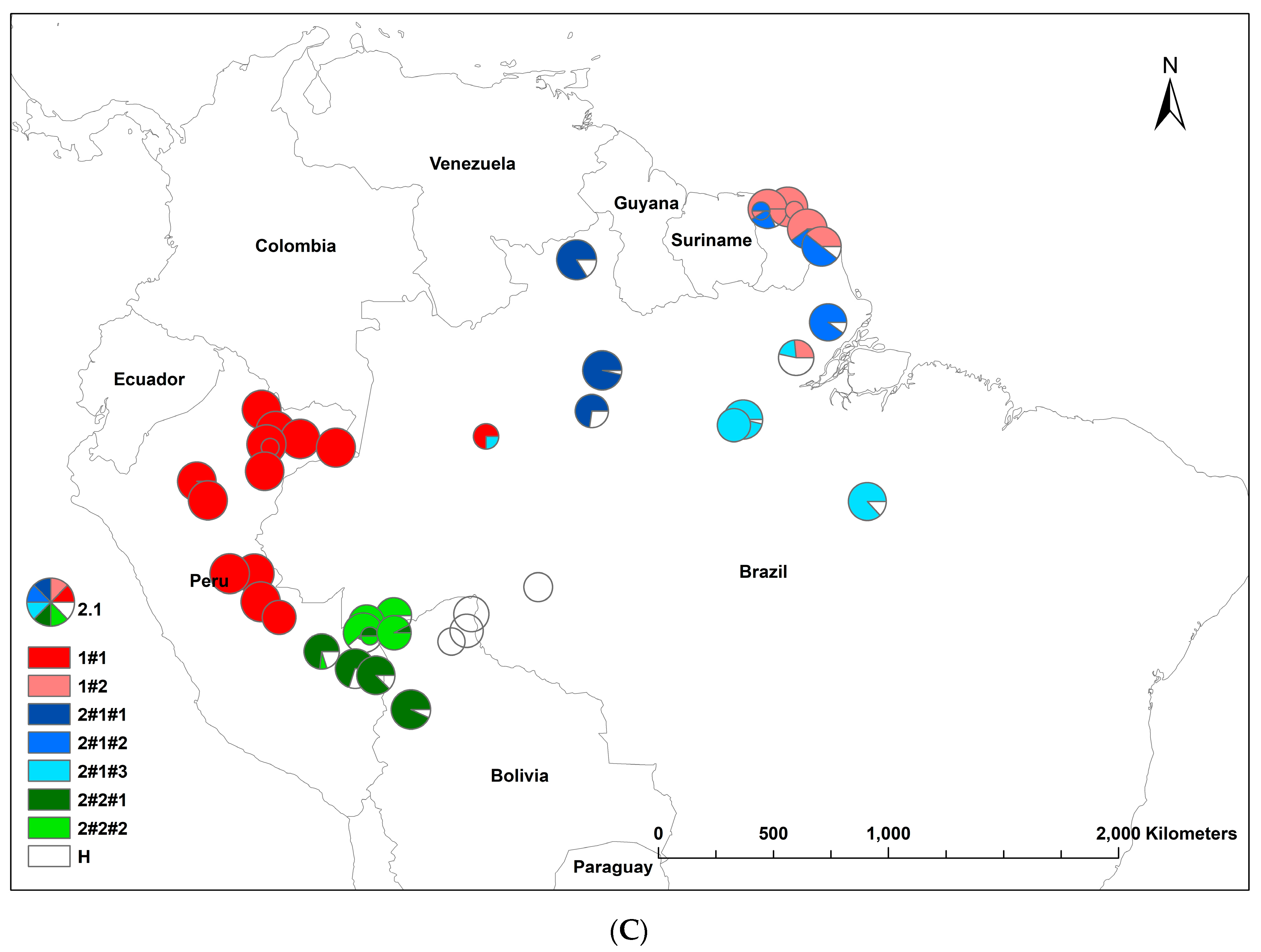
| Country | Sampling Site/Population Sample | n | Latitude | Longitude | Abbrev | n1 |
|---|---|---|---|---|---|---|
| 1-F. Guiana | Counami | 30 | 5.41543 | −53.175 | 1FG-Co | 32 |
| 2-F. Guiana | Sinnamary | 2 | 5.2884 | −52.916 | ||
| 3-F. Guiana | Piste de Paul Isnard | 27 | 5.33216 | −53.957 | 2FG-Is | 29 |
| 4-F. Guiana | Apatou | 2 | 5.27343 | −54.218 | ||
| 5-F. Guiana | Route de Cacao | 30 | 4.56779 | −52.406 | 3FG-Ro | 32 |
| 6-F. Guiana | Regina | 2 | 4.13118 | −52.088 | ||
| 7-F. Guiana | Saut Maripa | 28 | 3.87833 | −51.857 | 4FG-Ma | 28 |
| 8-Brazil | ESEC de Maraca-RR | 31 | 3.37032 | −61.444 | 5BW-Ma | 31 |
| 9-Brazil | Flona de Anauá e arredores-Rorainópolis-RR | 28 | −0.9339 | −60.451 | 6BW-An | 28 |
| 10-Brazil | AMATA Flona do Jamari-RO | 8 | −9.4014 | −62.911 | 7BW-Ja | 8 |
| 11-Brazil | ESEC do Jarí | 15 | −0.4955 | −52.829 | 8BW-Jr | 15 |
| 12-Brazil | Resex Chico Mendes-Xapuri-AC (AMATA-Flona do Jamari) | 16 | −10.504 | −68.595 | 9BW-Xa | 16 |
| 13-Brazil | Resex Chico Mendes-Comunidade Cumaru-Assis-AC | 15 | −10.772 | −69.647 | 10BW-Co | 15 |
| 14-Brazil | FLONA Amapá-AP | 20 | 0.52785 | −51.128 | 11BE-Am | 20 |
| 15-Brazil | PARNA da Ana Avilhanas-AM | 11 | −2.5345 | −60.837 | 12BE-Av | 11 |
| 16-Brazil | Flona de Tapajós-PA | 27 | −2.8687 | −54.92 | 13BE-Ta | 27 |
| 17-Brazil | Resex Tapajós-Arapins-PA | 11 | −3.0792 | −55.278 | 14BE-Ar | 11 |
| 18-Brazil | FLONA Tefé-AM | 4 | −3.5248 | −64.972 | 15BE-Te | 4 |
| 19-Brazil | FLONA do Carajás | 23 | −6.0628 | −50.059 | 16BE-Ca | 23 |
| 20-Peru | Dpto Loreto, Maynas, El Napo, Huiririma Native Community | 26 | −2.4761 | −73.744 | 17PN-Hu | 26 |
| 21-Peru | Huaman Urco | 27 | −3.3128 | −73.198 | 18PN-Ur | 27 |
| 22-Peru | Dpto Loreto, Maynas, Las Amazonas, Est. Biológica Madreselva | 28 | −3.6312 | −72.233 | 19PN-Ma | 28 |
| 23-Peru | Dpto Loreto, Mayna, Iquitos, Comunidad Campesina Yarina | 28 | −3.827 | −73.567 | 20PN-Ya | 28 |
| 24-Peru | Allpahuayo | 2 | −3.9544 | −73.422 | ||
| 25-Peru | Dpto Loreto, Mar. Ramón Castilla, C. Poblado Unión Progresista | 27 | −3.9727 | −70.841 | 21PN-Pr | 29 |
| 26-Peru | Dpto Loreto, Requena, Jenaro Herrera Research Centre | 11 | −4.8966 | −73.646 | 22PN-Re | 11 |
| 27-Peru | Jenaro Herrera | 25 | −4.9158 | −73.649 | 23PN-He | 25 |
| 28-Peru | Dpto Loreto, Alto Amazonas, Jeberos, Centro Poblado Jeberos | 26 | −5.2598 | −76.317 | 24PN-Je | 26 |
| 29-Peru | Shucushuyacu | 27 | −6.0199 | −75.854 | 25PN-Sh | 27 |
| 30-Peru | Dpto Ucayali, Cor. Portillo, Con. Forestal-Oxigeno para el Mundo | 29 | −8.8869 | −74.034 | 26PS-Po | 29 |
| 31-Peru | Dpto Ucayali, Padre Abad, Macuya Forestry Research Station | 30 | −8.8766 | −75.014 | 27PS-Pa | 30 |
| 32-Peru | Dpto Ucayali, Atalaya, Tahuania, Concesión Forestal-Javier Díaz | 29 | −9.9803 | −73.817 | 28PS-Di | 29 |
| 33-Peru | Dpto Ucayali, Atalaya, Raymondi, Comunidad San Juan de Inuya | 12 | −10.582 | −73.071 | 29PS-In | 12 |
| 34-Peru | Dpto Madre de Dios, Tahuamanu, Concesión Forestal Maderacre | 31 | −11.145 | −69.758 | 30PS-Md | 33 |
| 35-Peru | Ibéria | 2 | −11.299 | −69.524 | ||
| 36-Peru | Dpto Madre de Dios, P.N. Manu, Est. Biológica Cocha Cashu | 15 | −11.903 | −71.403 | 31PS-Ca | 15 |
| 37-Peru | Dpto Madre de Dios, Manu, Estación Biológica Los Amigos | 30 | −12.565 | −70.088 | 32PS-Am | 30 |
| 38-Peru | Dpto Madre de Dios, R. Nac. Tambopata, La Torre-Sandoval | 24 | −12.832 | −69.284 | 33PS-Ta | 24 |
| 39-Bolivia | Riberalta, MABET | 15 | −10.442 | −65.55 | 34Bo-Ri | 15 |
| 40-Bolivia | Riberalta, El Desvelo | 11 | −11.093 | −65.746 | 35Bo-De | 11 |
| 41-Bolivia | Cobija, Road—Bella Vista | 13 | −11.198 | −68.287 | 36Bo-Vi | 13 |
| 42-Bolivia | Riberalta, El Chorro | 5 | −11.514 | −66.327 | 37Bo-Ch | 5 |
| 43-Bolivia | Rurrenabaque, Área Protegida Madidi | 29 | −14.162 | −67.905 | 38Bo-Ma | 29 |
| Sample | n | nSNPs | CpMtSNPs | nCpMtSNPs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1FG-Co | 32 | 6.6 | 122 | 50.4 | 0.023 | 0.028 | 0.005 | 16 | 6.7 | 45.3 |
| 2FG-Is | 29 | 4.9 | 190 | 69.0 | 0.109 | 0.197 | 0.25 * | 17 | 13.3 | 62.5 |
| 3FG-Ro | 32 | 5.7 | 196 | 71.7 | 0.094 | 0.202 | 0.295 * | 16 | 6.7 | 64.1 |
| 4FG-Ma | 28 | 3.5 | 195 | 72.6 | 0.148 | 0.254 | 0.286 * | 16 | 13.3 | 65.7 |
| 5BW-Ma | 31 | 2.4 | 193 | 82.3 | 0.299 | 0.309 | 0.005 | 16 | 6.7 | 73.4 |
| 6BW-An | 28 | 1.5 | 195 | 70.8 | 0.291 | 0.292 | −0.022 | 15 | 0 | 72.7 |
| 7BW-Ja | 8 | 32.3 | 207 | 80.5 | 0.194 | 0.298 | 0.198 * | 17 | 13.3 | 72.6 |
| 8BW-Jr | 15 | 4.3 | 207 | 83.2 | 0.205 | 0.287 | 0.057 | 16 | 13.3 | 75.0 |
| 9BW-Xa | 16 | 4.5 | 204 | 72.6 | 0.278 | 0.290 | −0.021 | 16 | 6.7 | 75.0 |
| 10BW-Cu | 15 | 1.8 | 147 | 83.2 | 0.261 | 0.275 | 0.016 | 15 | 0 | 73.5 |
| 11BE-Am | 20 | 1.1 | 204 | 75.2 | 0.215 | 0.231 | 0.052 | 15 | 0 | 66.4 |
| 12BE-Av | 11 | 7.5 | 207 | 61.9 | 0.278 | 0.290 | −0.021 | 15 | 0 | 64.8 |
| 13BE-Ta | 27 | 2.8 | 197 | 83.2 | 0.261 | 0.275 | 0.016 | 15 | 0 | 74.2 |
| 14BE-Ar | 11 | 3.1 | 183 | 69.0 | 0.238 | 0.276 | 0.078 | 15 | 0 | 68.0 |
| 15BE-Te | 4 | 1.4 | 207 | 83.2 | 0.141 | 0.257 | 0.057 | 15 | 0 | 73.5 |
| 16BE-Ca | 23 | 5.3 | 190 | 81.4 | 0.275 | 0.282 | −0.019 | 15 | 0 | 75.0 |
| 17PN-Hu | 26 | 2.4 | 149 | 30.1 | 0.053 | 0.059 | 0.017 | 17 | 13.3 | 28.1 |
| 18PN-Ur | 27 | 5.0 | 147 | 31.9 | 0.060 | 0.066 | 0.018 | 17 | 13.3 | 29.7 |
| 19PN-Ma | 28 | 4.4 | 147 | 30.1 | 0.054 | 0.066 | 0.031 | 19 | 20.0 | 28.9 |
| 20PN-Ya | 28 | 2.5 | 147 | 30.1 | 0.059 | 0.064 | 0.024 | 16 | 6.7 | 27.4 |
| 21PN-Pr | 29 | 4.7 | 142 | 30.1 | 0.065 | 0.069 | 0.016 | 19 | 26.7 | 29.7 |
| 22PN-Re | 11 | 1.8 | 146 | 25.7 | 0.043 | 0.065 | 0.072 | 15 | 0 | 22.7 |
| 23PN-He | 25 | 3.8 | 145 | 29.2 | 0.050 | 0.056 | 0.018 | 16 | 20 | 28.1 |
| 24PN-Je | 26 | 7.9 | 142 | 28.3 | 0.041 | 0.061 | 0.087 * | 19 | 26.7 | 28.1 |
| 25PN-Sh | 27 | 3.2 | 147 | 26.5 | 0.047 | 0.053 | 0.028 | 14 | 0 | 23.4 |
| 26PS-Po | 29 | 13.9 | 146 | 30.1 | 0.046 | 0.075 | 0.087 * | 16 | 6.7 | 27.4 |
| 27PS-Pa | 30 | 3.1 | 140 | 30.1 | 0.063 | 0.064 | 0.020 | 14 | 0 | 26.6 |
| 28PS-Di | 29 | 4.9 | 139 | 23.9 | 0.054 | 0.061 | 0.024 | 15 | 0 | 21.1 |
| 29PS-In | 12 | 4.8 | 213 | 23.0 | 0.056 | 0.061 | 0.017 | 15 | 0 | 20.3 |
| 30PS-Ma | 33 | 4.5 | 206 | 88.5 | 0.297 | 0.315 | 0.045 | 15 | 0 | 78.1 |
| 31PS-Ca | 15 | 2.4 | 213 | 82.3 | 0.271 | 0.310 | 0.073 | 15 | 0 | 72.7 |
| 32PS-Am | 30 | 4.5 | 209 | 88.5 | 0.312 | 0.317 | −0.010 | 15 | 0 | 78.1 |
| 33PS-Ta | 24 | 3.2 | 209 | 85.0 | 0.284 | 0.314 | 0.080 | 15 | 0 | 75.0 |
| 34Bo-Mb | 15 | 2.8 | 207 | 84.1 | 0.321 | 0.324 | −0.026 | 16 | 6.7 | 75.0 |
| 35Bo-De | 11 | 1.3 | 206 | 84.1 | 0.297 | 0.312 | −0.012 | 16 | 6.7 | 75.0 |
| 36Bo-Vi | 13 | 0.8 | 205 | 82.3 | 0.313 | 0.313 | −0.023 | 15 | 0 | 72.7 |
| 37Bo-Ch | 5 | 1.4 | 212 | 82.3 | 0.340 | 0.348 | −0.086 | 19 | 26.7 | 75.8 |
| 38Bo-Ma | 29 | 1.3 | 226 | 87.6 | 0.322 | 0.313 | −0.032 | 15 | 0 | 77.3 |
| Overall | 832 | 4.4 | 183 | 100 | 0.178 | 0.204 | 0.086 * | 15.9 | 6.7 | 55.9 |
| French Guiana | 121 | 5.2 | 200 | 76.1 | 0.095 | 0.192 | 0.506 * | 18 | 20.0 | 69.5 |
| Brazil | 209 | 4.2 | 226 | 100 | 0.261 | 0.354 | 0.264 * | 30 | 100 | 100 |
| Peru | 429 | 4.7 | 217 | 92.9 | 0.111 | 0.222 | 0.498 * | 23 | 53.3 | 88.3 |
| Bolivia | 73 | 1.5 | 218 | 92.9 | 0.319 | 0.359 | 0.113 * | 20 | 33.3 | 85.9 |
| Sample | nCpMtSNPs (128) | nSNPs (113) | CpMtSNPs (15) | |
|---|---|---|---|---|
| All population samples | 38 | 0.484 ± 0.043 * | 0.415 ± 0.032 * | 0.942 ± 0.042 * |
| Genetic groups | 8, 9, and 4 | 0.401 ± 0.044 * | 0.315 ± 0.03 * | 0.896 ± 0.094 * |
| Countries | 4 | 0.295 ± 0.036 * | 0.233 ± 0.022 * | 0.695 ± 0.144 * |
| French Guiana | 4 | 0.120 ± 0.017 * | 0.117 ± 0.017 * | 0.011 ± 0.002 |
| Brazil | 12 | 0.299 ± 0.049 * | 0.224 ± 0.03 * | 0.925 ± 0.103 * |
| Peru | 17 | 0.466 ± 0.056 * | 0.456 ± 0.056 * | 0.741 ± 0.267 * |
| Bolivia | 5 | 0.142 ± 0.034 * | 0.107 ± 0.024 * | 0.735 ± 0.383 * |
| Reporting Groups | Self-Assignment Test of Grouped Individuals | Self-Assignment Test of Individuals | ||||
|---|---|---|---|---|---|---|
| Rate | Score | Rate | Score | Rate: Score > 80% | Rate: Score > 95% | |
| 1 | 100 | 100 | 100 | 99.9 | 100 | 99.4 |
| 2 | 100 | 100 | 100 | 100 | 100 | 100 |
| 3 | 100 | 100 | 98.6 | 99.8 | 100 | 98.6 |
| 4 | 100 | 100 | 96.0 | 93.2 | 89.7 | 69.2 |
| 5 | 100 | 100 | 100 | 100 | 100 | 100 |
| 6 | 100 | 100 | 94.5 | 91.7 | 82.6 | 67.4 |
| 7 | 100 | 100 | 100 | 98.2 | 96.5 | 94.7 |
| Overall | 100 | 100 | 98.4 | 97.5 | 95.5 | 89.9 |
| Country | ||||||
| French Guiana | 100 | 100 | 87.6 | 98.1 | 99.1 | 99.1 |
| Brazil | 100 | 100 | 98.1 | 99.9 | 100 | 98.5 |
| Peru | 100 | 100 | 98.1 | 93.4 | 87.8 | 72.8 |
| Bolivia | 100 | 100 | 100 | 99.9 | 100 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capo, L.F.M.; Degen, B.; Blanc-Jolivet, C.; Tysklind, N.; Cavers, S.; Mader, M.; Meyer-Sand, B.R.V.; Paredes-Villanueva, K.; Honorio Conorado, E.N.; García-Dávila, C.R.; et al. Timber Tracking of Jacaranda copaia from the Amazon Forest Using DNA Fingerprinting. Forests 2024, 15, 1478. https://doi.org/10.3390/f15081478
Capo LFM, Degen B, Blanc-Jolivet C, Tysklind N, Cavers S, Mader M, Meyer-Sand BRV, Paredes-Villanueva K, Honorio Conorado EN, García-Dávila CR, et al. Timber Tracking of Jacaranda copaia from the Amazon Forest Using DNA Fingerprinting. Forests. 2024; 15(8):1478. https://doi.org/10.3390/f15081478
Chicago/Turabian StyleCapo, Lorena Frigini Moro, Bernd Degen, Celine Blanc-Jolivet, Niklas Tysklind, Stephen Cavers, Malte Mader, Barbara Rocha Venancio Meyer-Sand, Kathelyn Paredes-Villanueva, Eurídice Nora Honorio Conorado, Carmen Rosa García-Dávila, and et al. 2024. "Timber Tracking of Jacaranda copaia from the Amazon Forest Using DNA Fingerprinting" Forests 15, no. 8: 1478. https://doi.org/10.3390/f15081478
APA StyleCapo, L. F. M., Degen, B., Blanc-Jolivet, C., Tysklind, N., Cavers, S., Mader, M., Meyer-Sand, B. R. V., Paredes-Villanueva, K., Honorio Conorado, E. N., García-Dávila, C. R., Troispoux, V., Delcamp, A., & Sebbenn, A. M. (2024). Timber Tracking of Jacaranda copaia from the Amazon Forest Using DNA Fingerprinting. Forests, 15(8), 1478. https://doi.org/10.3390/f15081478






