Integrated Metabolome and Transcriptome Analysis Reveals New Insights into the Walnut Seed Coat Coloration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sample Preparation
2.3. Determination of Total Anthocyanin Content by Spectrophotometry
2.4. Anthocyanin Targeted Metabolomics Analysis
2.5. Transcriptomic Analysis
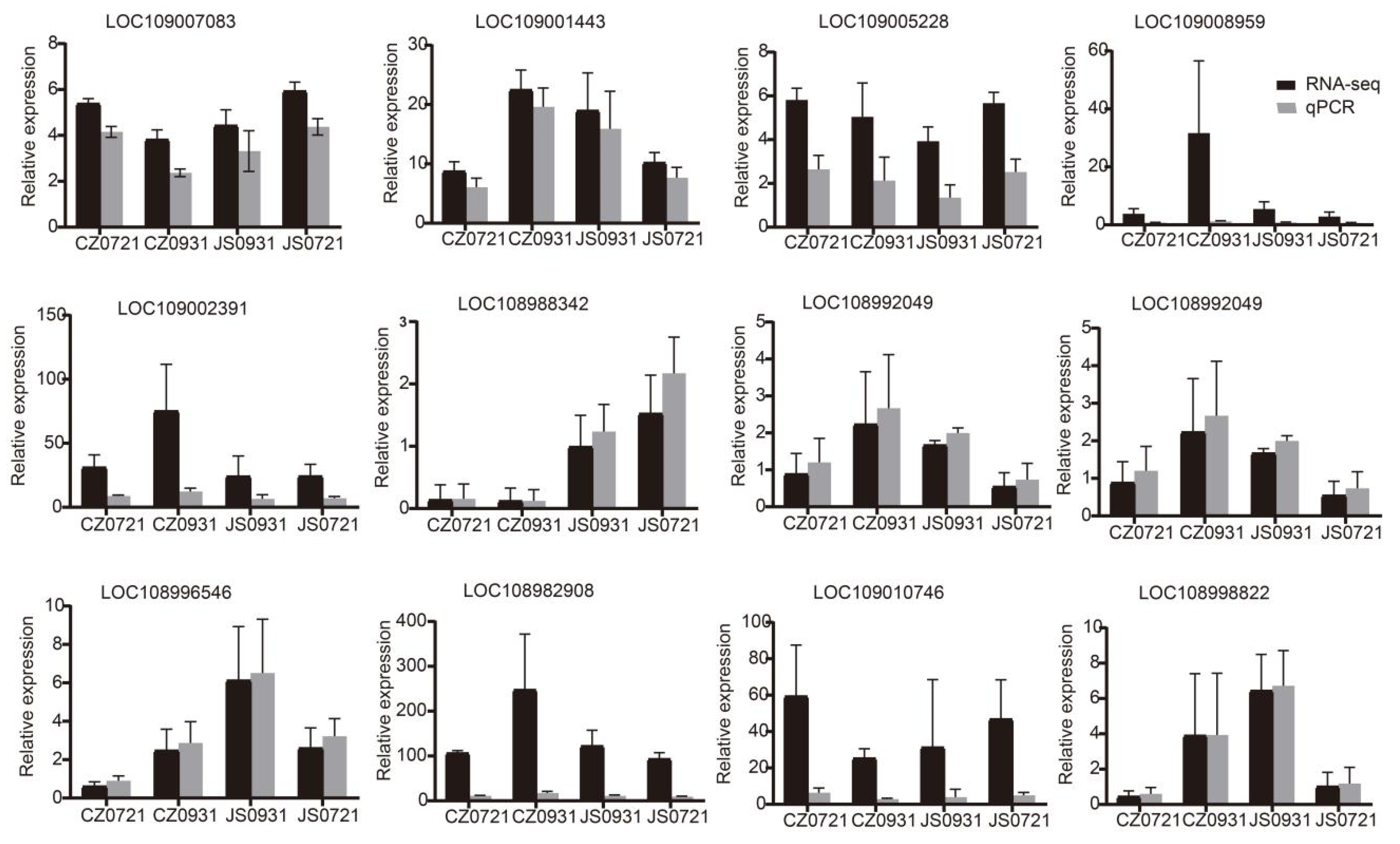
2.6. RT-qPCR Analysis
3. Results
3.1. Morphology and Anthocyanin Content of the Seed Color Development
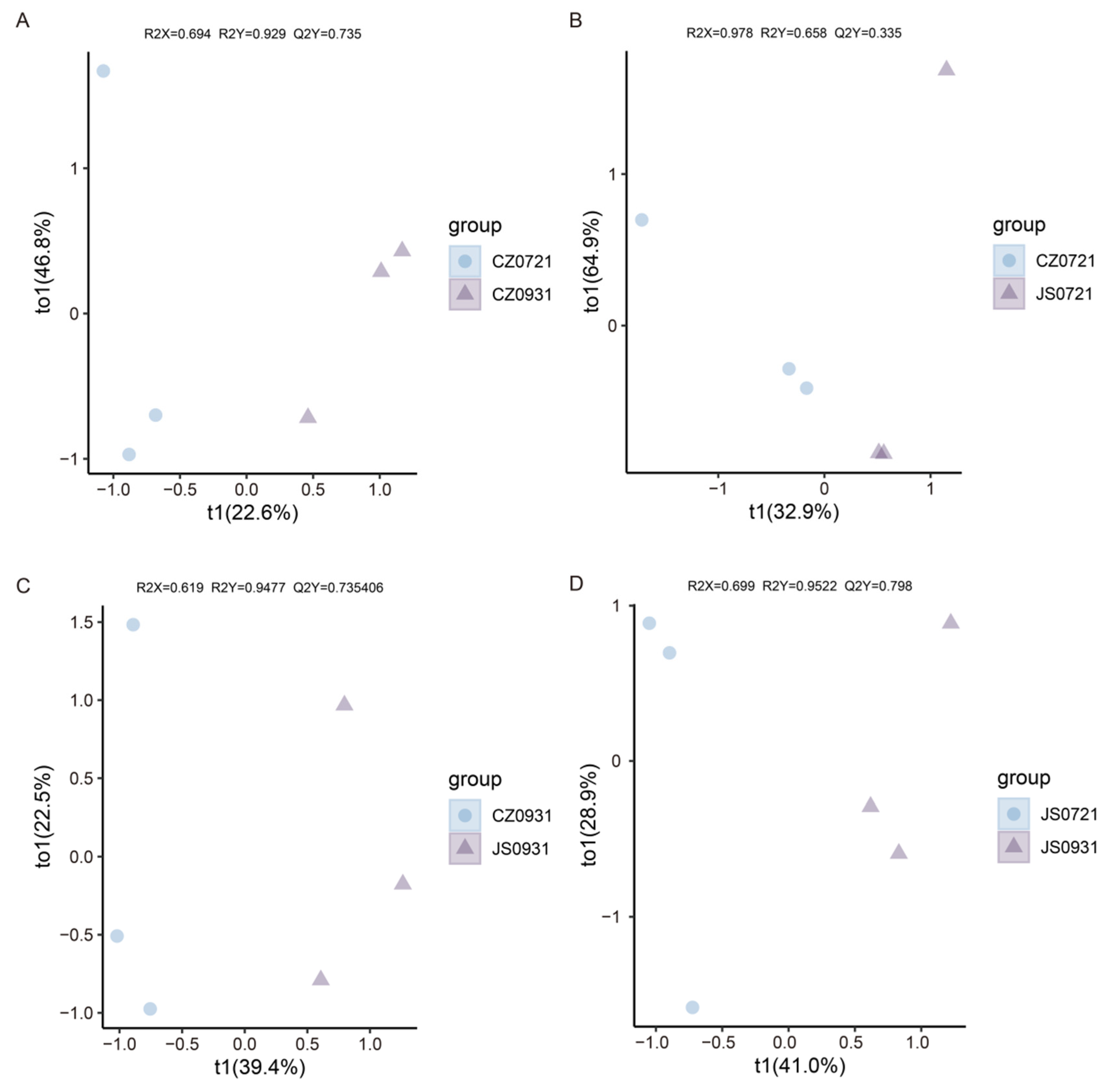
3.2. Comparison of Metabolites Among Clones at Different Developmental Stages
3.3. Transcriptome Sequencing and Differential Gene Expression Analysis
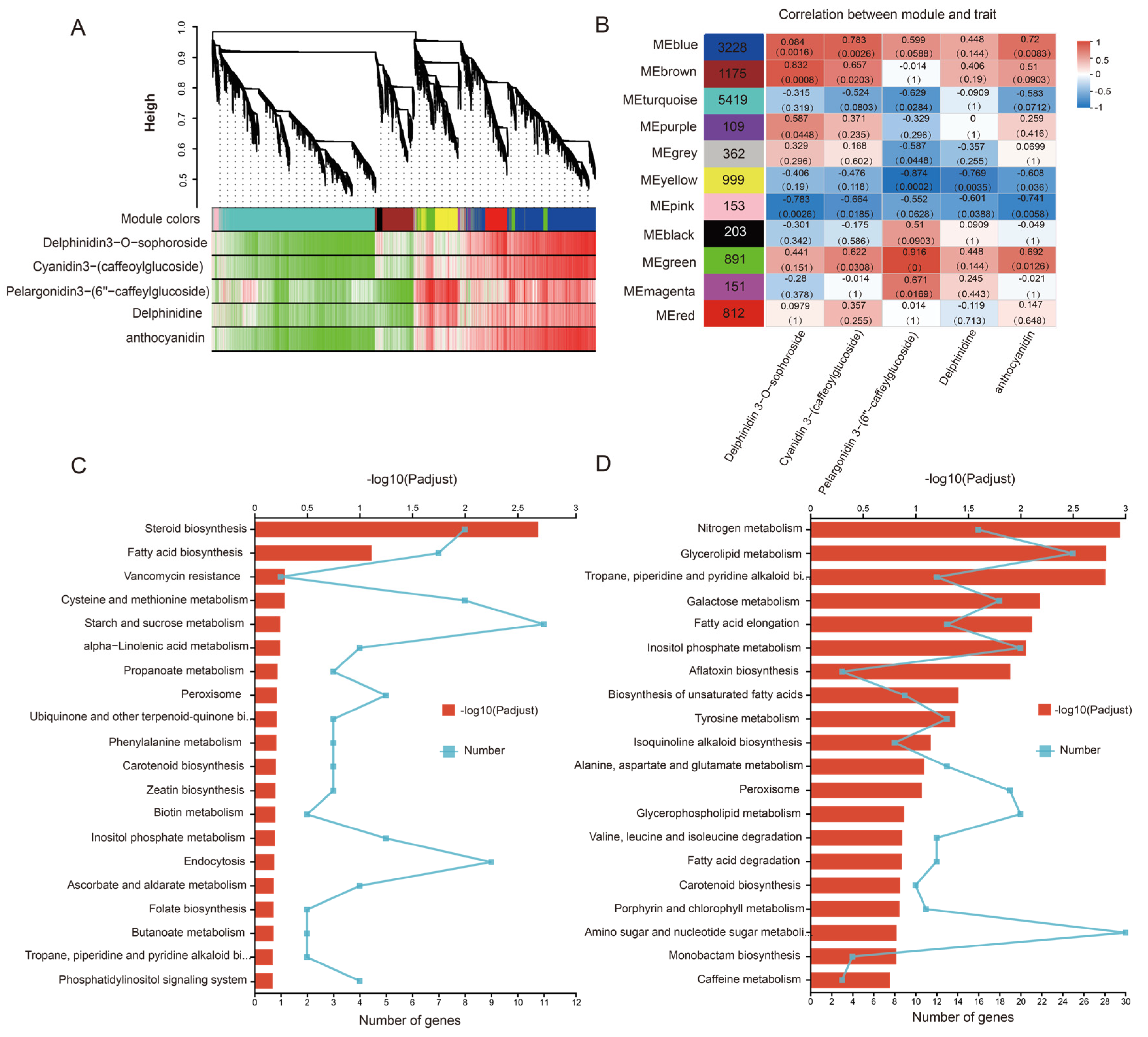
3.4. Weighted Gene Coexpression Network Analysis (WGCNA)
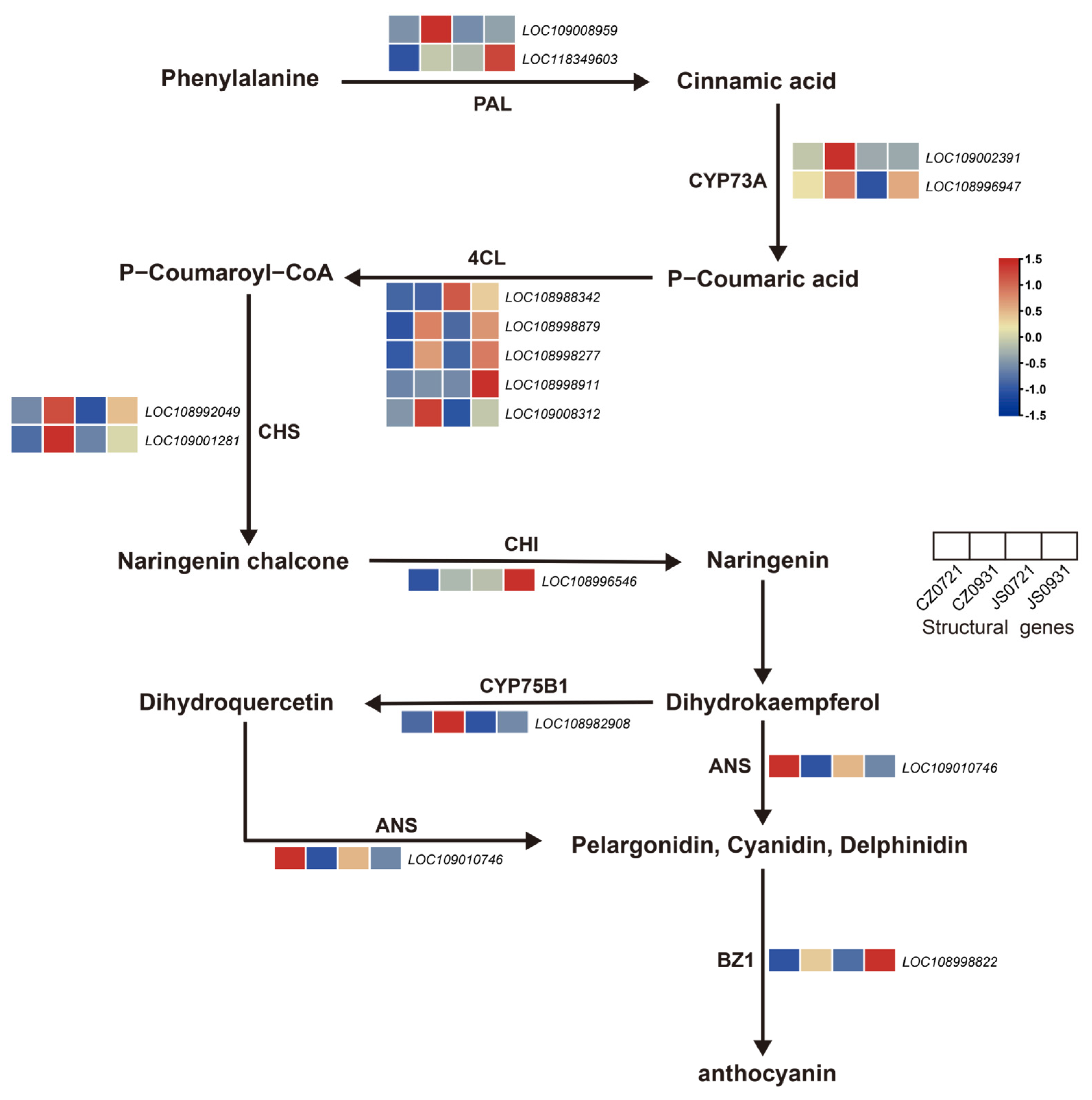
3.5. Anthocyanin Pathway of Seed Coat Coloration
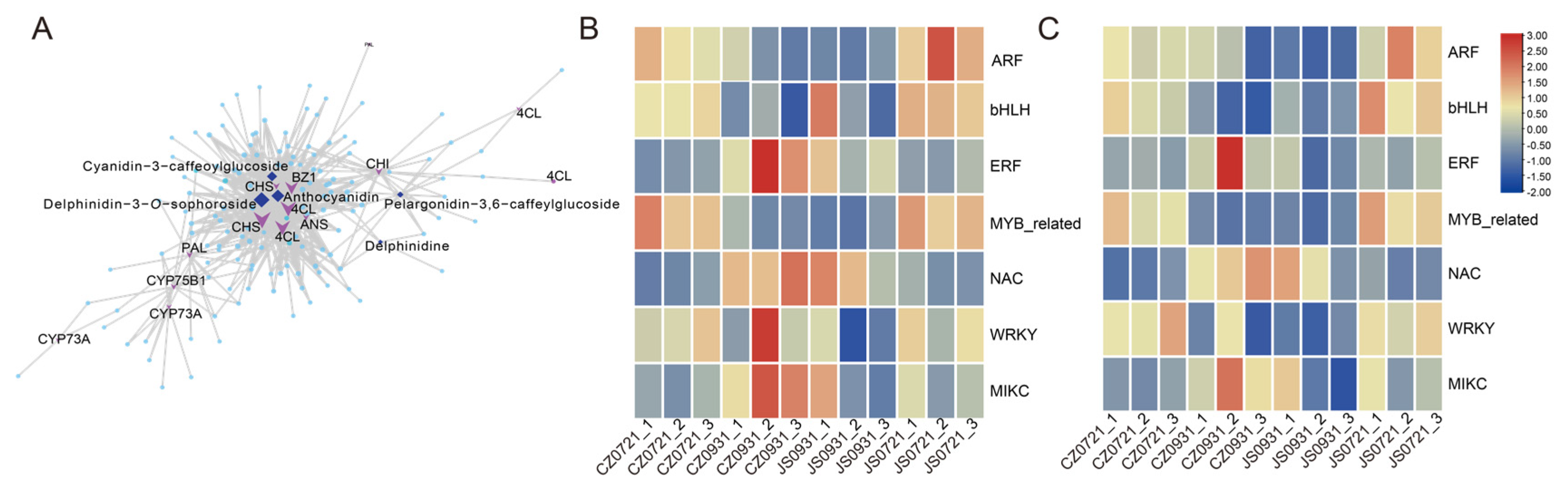
3.6. Transcriptional Regulatory Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, M.; Li, C.; Cao, C.; Wang, L.; Li, X.; Che, J.; Yang, H.; Zhang, X.; Zhao, H.; He, G.; et al. Walnut Fruit Processing Equipment: Academic Insights and Perspectives. Food Eng. Rev. 2021, 13, 822–857. [Google Scholar] [CrossRef]
- Li, Z.; Yang, L.; Abuliz, B.; Basti, A.; Yan, S.; Liu, K.; Zhu, Z. Research on the development of walnut industry in China. Chin. J. Agric. Mech. 2013, 34, 23–28. [Google Scholar] [CrossRef]
- Marrano, A.; Sideli, G.M.; Leslie, C.A.; Cheng, H.; Neale, D.B. Deciphering of the Genetic Control of Phenology, Yield, and Pellicle Color in Persian Walnut (Juglans regia L.). Front. Plant Sci. 2019, 10, 1140. [Google Scholar] [CrossRef]
- Fuentealba, C.; Hernández, I.; Saa, S.; Toledo, L.; Burdiles, P.; Chirinos, R.; Campos, D.; Brown, P.; Pedreschi, R. Colour and in vitro quality attributes of walnuts from different growing conditions correlate with key precursors of primary and secondary metabolism. Food Chem. 2017, 232, 664–672. [Google Scholar] [CrossRef]
- Li, Y.; Luo, X.; Wu, C.; Cao, S.; Zhou, Y.; Jie, B.; Cao, Y.; Meng, H.; Wu, G. Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in Red and Green Walnut (Juglans regia L.). Molecules 2017, 23, 25. [Google Scholar] [CrossRef]
- Ma, H.; Yang, T.; Li, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell 2021, 33, 3309–3330. [Google Scholar] [CrossRef]
- Huang, H.; Zhao, L.; Zhang, B.; Huang, W.; Zhang, Z.; An, B. Integrated analysis of the metabolome and transcriptome provides insights into anthocyanin biosynthesis of cashew apple. Food Res. Int. 2024, 175, 113711. [Google Scholar] [CrossRef]
- Han, Y.; Lu, M.; Yue, S.; Li, K.; Shang, F. Transcriptome and metabolome profiling revealing anthocyanin and phenolic acid biosynthetic mechanisms in sweet osmanthus pericarp. Sci. Hortic. 2021, 289, 110489. [Google Scholar] [CrossRef]
- Wu, Y.; Han, T.; Lyu, L.; Li, W.; Wu, W. Research progress in understanding the biosynthesis and regulation of plant anthocyanins. Sci. Hortic. 2023, 321, 112374. [Google Scholar] [CrossRef]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef]
- Sunil, L.; Shetty, N.P. Biosynthesis and regulation of anthocyanin pathway genes. Appl. Microbiol. Biotechnol. 2022, 106, 1783–1798. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, Z.; Sun, X. Single-cell and spatial multi-omics in the plant sciences: Technical advances, applications, and perspectives. Plant Commun. 2023, 4, 100508. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Feng, Y.; Zhou, S.; Zhang, J.; Guo, B.; Xiong, Y.; Wu, S.; Li, Y.; Li, Y.; Li, C. Metabolomics and transcriptomics provide insights into the molecular mechanisms of anthocyanin accumulation in the seed coat of differently colored mung bean (Vigna radiata L.). Plant Physiol. Biochem. 2023, 200, 107739. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; He, S.; Liu, Y.; Liu, B.; Ju, Y.; Kang, D.; Sun, X.; Fang, Y. Transcriptomics integrated with metabolomics reveals the effect of regulated deficit irrigation on anthocyanin biosynthesis in Cabernet Sauvignon grape berries. Food Chem. 2020, 314, 126170. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Ouyang, L.; Yao, R.; He, D.; Han, Z.; Li, W.; Ding, Y.; Wang, Z.; Kang, Y.; et al. Metabolomics combined with transcriptomics analyses of mechanism regulating testa pigmentation in peanut. Front. Plant Sci. 2022, 13, 1065049. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Li, P.; Sun, C.; Dong, W. Integrative metabolome and transcriptome analyses reveals the black fruit coloring mechanism of Crataegus maximowiczii C. K. Schneid. Plant Physiol. Biochem. 2023, 194, 111–121. [Google Scholar] [CrossRef]
- Liu, B.; Zhu, J.; Lin, L.; Yang, Q.; Hu, B.; Wang, Q.; Zou, X.-X.; Zou, S.-Q. Genome-Wide Identification and Co-Expression Analysis of ARF and IAA Family Genes in Euscaphis konishii: Potential Regulators of Triterpenoids and Anthocyanin Biosynthesis. Front. Genet. 2022, 12, 737293. [Google Scholar] [CrossRef]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying Advanced Metabolomic Data Processing and Subsequent Statistical Analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef]
- Navarro-Reig, M.; Jaumot, J.; García-Reiriz, A.; Tauler, R. Evaluation of changes induced in rice metabolome by Cd and Cu exposure using LC-MS with XCMS and MCR-ALS data analysis strategies. Anal. Bioanal. Chem. 2015, 407, 8835–8847. [Google Scholar] [CrossRef]
- Gagnebin, Y.; Tonoli, D.; Lescuyer, P.; Ponte, B.; de Seigneux, S.; Martin, P.-Y.; Schappler, J.; Boccard, J.; Rudaz, S. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Anal. Chim. Acta 2017, 955, 27–35. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. Am. Chem. Soc. 2015, 14, 1535–3893. [Google Scholar] [CrossRef] [PubMed]
- Komivi, D.; Marie, A.M.; Rong, Z.; Qi, Z.; Mei, Y.; Ndiaga, C.; Diaga, D.; Linhai, W.; Xiurong, Z. The contrasting response to drought and waterlogging is underpinned by divergent DNA methylation programs associated with transcript accumulation in sesame. Plant Sci. 2018, 277, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Mmadi, M.A.; Zhou, R.; Zhang, T.; Su, R.; Zhang, Y.; Wang, L.; You, J.; Zhang, X. Depicting the Core Transcriptome Modulating Multiple Abiotic Stresses Responses in Sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2019, 20, 3930. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Ma, T.; Sun, X.; Zhao, J.; You, Y.; Lei, Y.; Gao, G.; Zhan, J. Corrigendum to “Nutrient Compositions and Antioxidant Capacity of Kiwifruit (Actinidia) and their relationship with flesh color and commercial value” [Food Chem. 218 (2017) 294–304]. Food Chem. 2017, 224, 439. [Google Scholar] [CrossRef]
- Iglesias, I.; Alegre, S. The Effects of Reflective Film on Fruit Color, Quality, Canopy Light Distribution, and Profitability of ‘Mondial Gala’ Apples. HortTechnology 2024, 14, 488–498. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, K.; Ren, H.; Chang, J.; Yao, X. Variation in pigments in pecan testa during kernel development and storage. Food Chem. 2024, 438, 137989. [Google Scholar] [CrossRef]
- Wang, Y.; He, W.; Wang, L.; Lan, Y.; Wu, M. TCP transcription factor identification in pecan (Carya illinoensis) and salt tolerance function analysis of CiTCP8. Sci. Hortic. 2024, 330, 113051. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, Y.; Jin, F.; Zhang, J.; Ji, F.; Bai, Y.; Pei, D. Metabolome and Transcriptome Profiling Unveil the Mechanisms of Polyphenol Synthesis in the Developing Endopleura of Walnut (Juglans regia L.). Int. J. Mol. Sci. 2022, 23, 6623. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Q.; Lang, L.; Jiang, C.; Wang, X.; Sun, H. Integrated metabolome and transcriptome analysis of the anthocyanin biosynthetic pathway in relation to color mutation in miniature roses. BMC Plant Biol. 2021, 21, 257. [Google Scholar] [CrossRef]
- Fernández-García, E.; Carvajal-Lérida, I.; Jarén-Galán, M.; Garrido-Fernández, J.; Pérez-Gálvez, A.; Hornero-Méndez, D. Carotenoids bioavailability from foods: From plant pigments to efficient biological activities. Food Res. Int. 2012, 46, 438–450. [Google Scholar] [CrossRef]
- Gao, Y.-F.; Zhao, D.-H.; Zhang, J.-Q.; Chen, J.-S.; Li, J.-L.; Weng, Z.; Rong, L.-P. De novo transcriptome sequencing and anthocyanin metabolite analysis reveals leaf color of Acer pseudosieboldianum in autumn. BMC Genom. 2021, 22, 383. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Brugliera, F.; Tao, G.-Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/Blue Chrysanthemums—Metabolic Engineering of the Anthocyanin Biosynthetic Pathway Results in Novel Petal Colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef]
- Peng, J.; Dong, X.; Xue, C.; Liu, Z.; Cao, F. Exploring the Molecular Mechanism of Blue Flower Color Formation in Hydrangea macrophylla cv. “Forever Summer”. Front. Plant Sci. 2021, 12, 585665. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, J.; Liu, Z.; Wang, J.; Yang, B.; Chen, W.; Ou, L.; Dai, X.; Zhang, Z.; Zou, X. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in pepper fruit (Capsicum annuum L.). Food Chem. 2020, 306, 125629. [Google Scholar] [CrossRef]
- Feng, X.; Yu, Q.; Li, B.; Kan, J. Comparative analysis of carotenoids and metabolite characteristics in discolored red pepper and normal red pepper based on non-targeted metabolomics. Lwt 2022, 153, 112398. [Google Scholar] [CrossRef]
- Yu, M.; Bai, M.; Chen, M.; Zhang, G.; Zhao, Y.; Ma, Q.; Yang, L.; Gu, C. Identification of bHLH transcription factors and screening of anthocyanin-related genes in Lagerstroemia indica. Genetica 2024, 152, 179–197. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, T.; Yuan, L.; Qiu, F.; Tang, Y.; Li, D.; Zhang, F.; Zeng, L.; Yang, C.; Nagdy, M.M.; et al. A Fruit-Expressed MYB Transcription Factor Regulates Anthocyanin Biosynthesis in Atropa belladonna. Int. J. Mol. Sci. 2024, 25, 4963. [Google Scholar] [CrossRef]
- Zhuo, M.-G.; Wang, T.-Y.; Huang, X.-M.; Hu, G.-B.; Zhou, B.-Y.; Wang, H.-C.; Abbas, F. ERF transcription factors govern anthocyanin biosynthesis in litchi pericarp by modulating the expression of anthocyanin biosynthesis genes. Sci. Hortic. 2024, 337, 113464. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genom. 2012, 13, 75–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, G.; Yang, J.; Shi, G.; Niu, Z.; Liu, H.; Xu, N.; Wang, L. Integrated metabolomics and transcriptomics reveal molecular mechanisms of corolla coloration in Rhododendron dauricum L. Plant Physiol. Biochem. 2024, 207, 108438. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qiu, J.; Ding, L.; Huang, M.; Huang, S.; Yang, G.; Yin, J. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 2017, 112, 335–345. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Huang, X.; Wan, X.; Zhang, S.; Luo, X.; Qian, J.; He, F.; Chen, L.; Zhang, F.; Yang, H. Integrated Metabolome and Transcriptome Analysis Reveals New Insights into the Walnut Seed Coat Coloration. Forests 2025, 16, 691. https://doi.org/10.3390/f16040691
Wang R, Huang X, Wan X, Zhang S, Luo X, Qian J, He F, Chen L, Zhang F, Yang H. Integrated Metabolome and Transcriptome Analysis Reveals New Insights into the Walnut Seed Coat Coloration. Forests. 2025; 16(4):691. https://doi.org/10.3390/f16040691
Chicago/Turabian StyleWang, Ruiqi, Xin Huang, Xueqin Wan, Shuaiying Zhang, Xiandan Luo, Jianghong Qian, Fang He, Lianghua Chen, Fan Zhang, and Hanbo Yang. 2025. "Integrated Metabolome and Transcriptome Analysis Reveals New Insights into the Walnut Seed Coat Coloration" Forests 16, no. 4: 691. https://doi.org/10.3390/f16040691
APA StyleWang, R., Huang, X., Wan, X., Zhang, S., Luo, X., Qian, J., He, F., Chen, L., Zhang, F., & Yang, H. (2025). Integrated Metabolome and Transcriptome Analysis Reveals New Insights into the Walnut Seed Coat Coloration. Forests, 16(4), 691. https://doi.org/10.3390/f16040691





