Genus-Specific Molecular Markers for In Vitro Detection of Corinectria Forest Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Samples
2.2. DNA Extraction
2.3. Design of Molecular Markers SPECIFIC to the Genus Corinectria
2.4. Validation of Molecular Markers
2.4.1. PCR Conditions
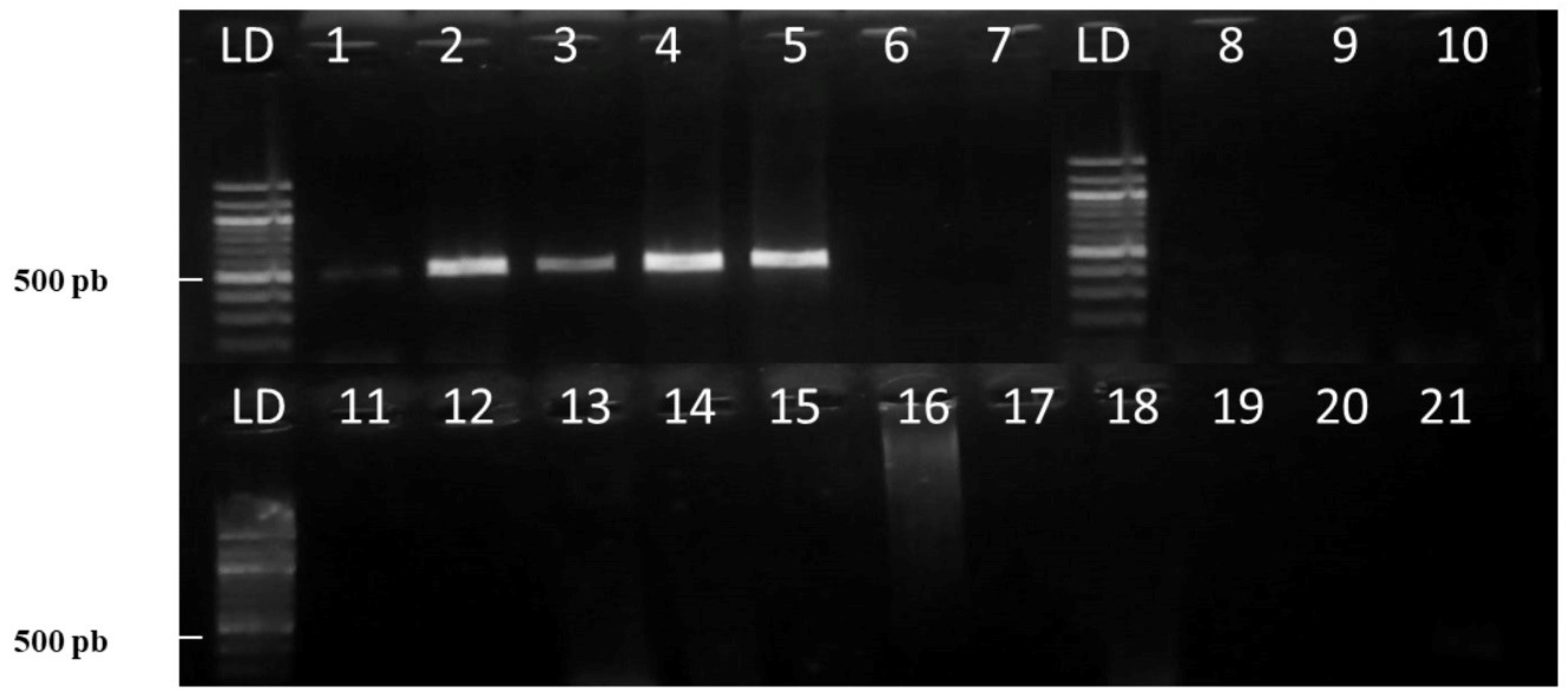
2.4.2. Marker Specificity
2.4.3. Marker Sensitivity
3. Results
3.1. Molecular Marker Design
3.1.1. Marker Specificity
3.1.2. Marker Sensitivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mead, D.J. Sustainable Management of Pinus Radiata Plantations; FAO Forestry Paper; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013; ISBN 978-92-5-107634-7. [Google Scholar]
- Ahumada, R.; Rotella, A.; Slippers, B.; Wingfield, M.J. Pathogenicity and Sporulation of Phytophthora pinifolia on Pinus radiata in Chile. Australas. Plant Pathol. 2013, 42, 413–420. [Google Scholar] [CrossRef]
- Morales, R. Detección de Neonectria fuckeliana en Chile, asociado a cancros y malformaciones fustales en plantaciones de Pinus radiata. Bosque 2009, 30, 106–110. [Google Scholar] [CrossRef]
- González, C.; Chaverri, P. Corinectria, a New Genus to Accommodate Neonectria fuckeliana and C. Constricta sp. nov. from Pinus radiata in Chile. Mycol. Prog. 2017, 16, 1015–1027. [Google Scholar] [CrossRef]
- Ahumada, R.; Rotella, A. Disease Management in the Forest Plantations in Chile. In Forest Pest and Disease Management in Latin America; Estay, S.A., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 171–184. ISBN 978-3-030-35142-7. [Google Scholar]
- González, C.; Morales, R.; Riegel, R.; Aravena, M.; Valenzuela, E. Distribución Geográfica y Caracterización Fenotípica y Molecular de Neonectria Fuckeliana, Asociado a Cancros Fustales de Pinus radiata En Chile. Bosque 2015, 36, 531–541. [Google Scholar] [CrossRef]
- González, C.; Morales, R.A.; Chaverri, P. Life Cycle and in Vitro Sporulation Dynamics of Corinectria constricta, the Causal Agent of Pinus radiata Stem Canker, in Chile. For. Path. 2020, 50, e12594. [Google Scholar] [CrossRef]
- Kulik, T.; Bilska, K.; Żelechowski, M. Promising Perspectives for Detection, Identification, and Quantification of Plant Pathogenic Fungi and Oomycetes through Targeting Mitochondrial DNA. IJMS 2020, 21, 2645. [Google Scholar] [CrossRef]
- Luchi, N.; Ioos, R.; Santini, A. Fast and Reliable Molecular Methods to Detect Fungal Pathogens in Woody Plants. Appl. Microbiol. Biotechnol. 2020, 104, 2453–2468. [Google Scholar] [CrossRef]
- Hariharan, G.; Prasannath, K. Recent Advances in Molecular Diagnostics of Fungal Plant Pathogens: A Mini Review. Front. Cell. Infect. Microbiol. 2021, 10, 600234. [Google Scholar] [CrossRef]
- Stewart, J.E.; Kim, M.-S.; Klopfenstein, N.B. Molecular Genetic Approaches Toward Understanding Forest-Associated Fungi and Their Interactive Roles Within Forest Ecosystems. Curr. For. Rep. 2018, 4, 72–84. [Google Scholar] [CrossRef]
- Brurberg, M.; Stensvand, A.; Talgø, V. Development and Application of a PCR-based Test for the Identification of Neonectria neomacrospora Damaging Abies Species. In Proceedings of the 12th International Christmas Tree Research and Extension Conference, Honne, Norway, 6–11 September 2015; p. 33. [Google Scholar]
- Langrell, S.R.H. Molecular Detection of Neonectria galligena (Syn. Nectria galligena). Mycol. Res. 2002, 106, 280–292. [Google Scholar] [CrossRef]
- Tripathi, A.; Dubey, S.C.; Akhtar, J.; Kumar, P. Development of PCR-Based Assays to Diagnose the Major Fungal Pathogens Infecting Pulse Crops, Potential for Germplasm Health Certification and Quarantine Processing. World J. Microbiol. Biotechnol. 2023, 39, 74. [Google Scholar] [CrossRef] [PubMed]
- Langrell, S.R. Development of a Nested PCR Detection Procedure for Nectria fuckeliana Direct from Norway Spruce Bark Extracts. FEMS Microbiol. Lett. 2005, 242, 185–193. [Google Scholar] [CrossRef]
- Santos, K.M.; Lima, G.S.; Barros, A.P.O.; Machado, A.R.; Souza-Motta, C.M.; Correia, K.C.; Michereff, S.J. Novel Specific Primers for Rapid Identification of Macrophomina Species. Eur. J. Plant Pathol. 2020, 156, 1213–1218. [Google Scholar] [CrossRef]
- Oliveira, M.; Azevedo, L. Molecular Markers: An Overview of Data Published for Fungi over the Last Ten Years. JoF 2022, 8, 803. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Luo, Y.; Zhou, S.; An, L.; Wang, C.; Jin, Q.; Zhou, M.; Xu, J.-R. Molecular Evolution and Functional Divergence of Tubulin Superfamily in the Fungal Tree of Life. Sci. Rep. 2014, 4, 6746. [Google Scholar] [CrossRef]
- Singh, N.; Kapoor, R. Quick and Accurate Detection of Fusarium oxysporum f. sp. carthami in Host Tissue and Soil Using Conventional and Real-Time PCR Assay. World J. Microbiol. Biotechnol. 2018, 34, 175. [Google Scholar] [CrossRef]
- Yeo, H.Y.; Dong, Y.S.; Seung, Y.S.; Seong, H.K. Development of PCR Method for Fast Detection of Ophiostoma floccosum in Wood Chips. Afr. J. Microbiol. Res. 2013, 7, 1913–1916. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.-J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K. Fusarium: More than a Node or a Foot-Shaped Basal Cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef]
- James, J.E.; Santhanam, J.; Zakaria, L.; Mamat Rusli, N.; Abu Bakar, M.; Suetrong, S.; Sakayaroj, J.; Abdul Razak, M.F.; Lamping, E.; Cannon, R.D. Morphology, Phenotype, and Molecular Identification of Clinical and Environmental Fusarium solani Species Complex Isolates from Malaysia. J. Fungi 2022, 8, 845. [Google Scholar] [CrossRef]
- Nielsen, K.N.; Thomsen, I.M.; Hansen, O.K. Direct Quantitative Real-Time PCR Assay for Detection of the Emerging Pathogen Neonectria neomacrospora. For. Pathol. 2019, 49, e12509. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for General Users and for Biologist Programmers. In Bioinformatics Methods and Protocols; Humana Press: Totowa, NJ, USA, 1999; Volume 132, pp. 365–386. ISBN 978-1-59259-192-3. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Chattopadhyay, A.; Tiwari, K.K.; Chaudhary, K.; Pratap, D. Genic Molecular Markers in Fungi: Availability and Utility for Bioprospection. Mol. Markers Mycol. Diagn. Marker Dev. 2017, 2017, 151–176. [Google Scholar]
- Olivieri, L.; Saville, R.J.; Gange, A.C.; Xu, X. Limited Asymptomatic Colonization of Apple Tree Shoots by Neonectria ditissima Following Infection of Leaf Scars and Pruning Wounds. Plant Pathol. 2021, 70, 1838–1849. [Google Scholar] [CrossRef]
- Capote, N.; Del Río, M.Á.; Herencia, J.F.; Arroyo, F.T. Molecular and Pathogenic Characterization of Cylindrocarpon-like Anamorphs Causing Root and Basal Rot of Almonds. Plants 2022, 11, 984. [Google Scholar] [CrossRef]
- Wallace, M.M.; Covert Sarah, F. Molecular Mating Type Assay for Fusarium circinatum. Appl. Environ. Microbiol. 2000, 66, 5506–5508. [Google Scholar] [CrossRef]
- Lombard, L.; Van Der Merwe, N.A.; Groenewald, J.Z.; Crous, P.W. Generic Concepts in Nectriaceae. Stud. Mycol. 2015, 80, 189–245. [Google Scholar] [CrossRef]
- Dizkirici, A.; Kalmer, A. Utility of Various Molecular Markers in Fungal Identification and Phylogeny. Nova Hedwig. 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Irimia, M.; Roy, S.W. Spliceosomal Introns as Tools for Genomic and Evolutionary Analysis. Nucleic Acids Res. 2008, 36, 1703–1712. [Google Scholar] [CrossRef]
- Msiska, Z.; Morton, J.B. Isolation and Sequence Analysis of a β-Tubulin Gene from Arbuscular Mycorrhizal Fungi. Mycorrhiza 2009, 19, 501–513. [Google Scholar] [CrossRef]


| Species | Isolate/Voucher a | Geographical Origen | Genbank Accession Number |
|---|---|---|---|
| Corinectria constricta | LASBE 330 | Chile | KY636422.1 |
| Corinectria constricta | LASBE 266 | Chile | KY636417.1 |
| Corinectria constricta | LASBE 352 | Chile | KY636425.1 |
| Corinectria fuckeliana | CBS 239.29 | Scotland | DQ789871 |
| Corinectria fuckeliana | AR 3103.61 | Austria | HM352857 |
| Phomopsis tuberívora | LSB 158 | Chile | - |
| Diaporthe phoenicicola | LSB 160 | Chile | - |
| Diplodia sapinea | LSB 161 | Chile | - |
| Fusarium verticillioides | LSB 162 | Chile | - |
| Neonectria ditissima | LSB 163 | Chile | - |
| Epicoccum nigrum | LSB 277 | Chile | - |
| Arthrinium sp. | LSB 278 | Chile | - |
| Trichoderma viride | LSB 279 | Chile | - |
| Oomycete | |||
| Phytophthora cinnamomi | LSB 159 | Chile | - |
| Plants | |||
| Pinus radiata | - | Chile | - |
| Species | Isolate/Voucher a | Genbank Accession Number | |
|---|---|---|---|
| ACT | Btub | ||
| Corinectria constricta | LASBE 260 | _ | KY636416.1 |
| Corinectria constricta | LASBE 266 | _ | KY636417.1 |
| Corinectria constricta | LASBE 284 | _ | KY636418.1 |
| Corinectria constricta | LASBE 301 | _ | KY636419.1 |
| Corinectria constricta | LASBE 306B | _ | KY636420.1 |
| Corinectria constricta | LASBE 314 | KY636431.1 | KY636421.1 |
| Corinectria constricta | LASBE 330 | KY636432.1 | KY636422.1 |
| Corinectria constricta | LASBE 340 | KY636433.1 | KY636423.1 |
| Corinectria constricta | LASBE 344 | KY636434.1 | KY636424.1 |
| Corinectria constricta | LASBE 352 | KY636435.1 | KY636425.1 |
| Neonectria fuckeliana | GJS02-67 | HM352886.1 | _ |
| Neonectria coccinea | CBS 119158; MAFF 241561 | KC660426.1 | DQ789892.1 |
| Neonectria faginata | CBS 134246; CBS 119198 | KC660414.1 | KC660743.1 |
| Neonectria microconidia | MAFF 241530; MAFF 241522 | KC660427.1 | KC660757.1 |
| Neonectria ditissima | CBS 100.316; CBS 226; CBS 100316; CBS 100318 | HM352880.1 | _ |
| Neonectria punicea | ABT12-1; CBS 119527; ABT12-1; CBS 124262; voucher 135 | KC660403.1; MW538899.1 | MW538902.1 |
| Name | Direction | Sequense (5-3 Prima) | Size | %GC a | Tm b |
|---|---|---|---|---|---|
| Act 81 F | forward | CGAGACTTTCAACGCCCC | 18 | 61.1 | 58.4 |
| Act 543 R | reverse | AGTGGTGACGTGAATGCC | 18 | 55.6 | 57.6 |
| Btub | forward | CTGATTCTACCCCGCCGAAG | 20 | 60 | 60.2 |
| BtubR | reverse | GCCAGAGGCCTAAGGGTTTT | 20 | 55 | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vásquez, T.; González, C.; Montalva, C. Genus-Specific Molecular Markers for In Vitro Detection of Corinectria Forest Pathogens. Forests 2025, 16, 697. https://doi.org/10.3390/f16040697
Vásquez T, González C, Montalva C. Genus-Specific Molecular Markers for In Vitro Detection of Corinectria Forest Pathogens. Forests. 2025; 16(4):697. https://doi.org/10.3390/f16040697
Chicago/Turabian StyleVásquez, Tania, Cristian González, and Cristian Montalva. 2025. "Genus-Specific Molecular Markers for In Vitro Detection of Corinectria Forest Pathogens" Forests 16, no. 4: 697. https://doi.org/10.3390/f16040697
APA StyleVásquez, T., González, C., & Montalva, C. (2025). Genus-Specific Molecular Markers for In Vitro Detection of Corinectria Forest Pathogens. Forests, 16(4), 697. https://doi.org/10.3390/f16040697








