Allometry for Biomass Estimation in Jatropha Trees Planted as Boundary Hedge in Farmers’ Fields
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
| Site (District) | Latitude | Longitude | Altitude (masl) | Mean annual Temperature (°C) | Rainfall (mm yr−1) |
|---|---|---|---|---|---|
| Salima | 13°45′0″S | 34°35′0″E | 512 | 24.0 | 1266 |
| Nkhotakota | 12°55′0″S | 34°17′0″E | 500 | 23.5 | 1649 |
| Dowa | 13°39′0″S | 33°55′6″E | 1514 | 19.3 | 906 |
| Kasungu | 13°02′0″S | 33°29′0″E | 1342 | 20.9 | 802 |
| Mzimba | 11°53′0″S | 33°37′0″E | 1349 | 19.6 | 903 |
2.2. Sampling and Tree Harvests
2.3. Data Analysis
| Tree variable | N | Min | Max | Mean | Std. dev. |
|---|---|---|---|---|---|
| Stratum 1 | |||||
| Wood biomass (kg) | 174 | 0.010 | 7.390 | 0.466 | 0.716 |
| Basal diameter (cm) | 174 | 1.4 | 12.8 | 3.770 | 1.504 |
| Height (m) | 174 | 0.05 | 3.16 | 0.823 | 0.494 |
| Stratum 2 | |||||
| Wood biomass (kg) | 442 | 0.006 | 2.616 | 0.168 | 0.256 |
| Basal diameter (cm) | 442 | 1.5 | 8.4 | 2.971 | 0.993 |
| Height (m) | 442 | 0.03 | 2.0 | 0.490 | 0.281 |
| Allometric model | |
|---|---|
| Log10 (wood biomass) = Log10 ( β0) + β1 × Log10 (BD) | I |
| Log10 (wood biomass) = Log10 ( β0) + β1 × Log10 (BD) + β1 × Log10 (Ht) | II |



3. Results
3.1. Model Specifications

| Model | Stratum | N | Log β0 | β1 | β2 | R2 | CF | Corrected equation |
|---|---|---|---|---|---|---|---|---|
| I | 1 | 172 | −2.196 | 2.891 | - | 0.726 | 1.039 | W = 0.0067 × BD2.891 |
| (0.077) | (0.136) | |||||||
| 2 | 442 | −2.281 | 2.769 | - | 0.556 | 1.046 | W = 0.0055 × BD2.769 | |
| (0.055) | (0.118) | |||||||
| II | 1 | 172 | −2.131 | 2.802 | 0.099 | 0.728 | ||
| (0.095) | (0.157) | (0.088) | ||||||
| 2 | 442 | −2.221 | 2.689 | 0.065 | 0.557 | |||
| (0.093) | (0.155) | (0.081) |
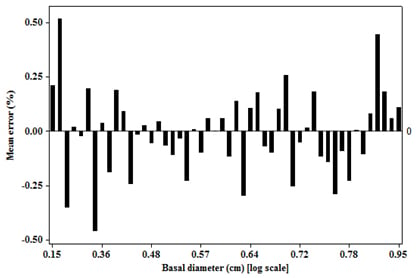
3.2. Residual Scatterplots

3.3. Goodness of Fit of the Model
| Model | Stratum | N | Bias MPE% 95% CI | Imprecision RMSE% 95% CI | ||
|---|---|---|---|---|---|---|
| 1 | 1 | 172 | 0.01 | −4.11 to 4.13 | 27.35 | 23.98 to 30.72 |
| 2 | 442 | −0.05 | −2.87 to 2.77 | 30.13 | 28.64 to 31.62 | |

3.4. Other Published Jatropha Models
| Model | Stratum | N | Bias MPE% 95% CI | Imprecision RMSE% 95% CI | ||
|---|---|---|---|---|---|---|
| Achten Model | 1 | 173 | −38.96 | −43.00 to −34.92 | 47.40 | 42.50 to 52.30 |
| Ghezehei model | 1 | 173 | −54.77 | −59.16 to −50.38 | 62.16 | 55.77 to 68.55 |
| Achten model | 2 | 293 | −20.02 | −23.41 to −16.63 | 35.64 | 32.29 to 38.99 |
| Ghezehei model | 2 | 293 | −45.14 | −48.77 to −41.51 | 55.09 | 50.56 to 59.62 |

4. Discussion
5. Conclusions
Acknowledgments
Conflict of Interest
References
- FAO, Forest Resource Assessment 2010; FAO: Rome, Italy, 2010.
- Government of Malawi, National Forest Act; Lilongwe, Malawi.
- Mascaro, J.; Litton, C.M.; Hughes, R.F.; Uowolo, A.; Schnitzer, S.A. Minimizing bias in biomass allometry: Model selection and log-transformation of data. Biotropica 2011, 43, 649–653. [Google Scholar] [CrossRef]
- Angelsen, A.; Brown, S.; Loisei, C.; Peskett, L.; Streck, C.; Zarin, D. Reducing Emissions from Deforestation and Forest Degradation (REDD); Technical Report for the Government of Norway; Meridian Institute: Washington, DC, USA, 2009. Available online: www.redd-oar.org (accessed on 7 November 2012).
- Preece, N.D.; Crowley, G.M.; Lawes, M.J.; van Oosterzee, P. Comparing aboveground biomass among forest types in wet tropics: Small stems and plantation types matter in carbon accounting. For. Ecol. Manag. 2012, 264, 228–237. [Google Scholar] [CrossRef]
- Chave, J.; Andalo, C.; Brown, S.; Cairns, M.A.; Chambers, J.Q.; Eamus, D.; Folster, H.; Fromard, F.; Higuchi, N.; Kira, T.; et al. Tree allometry and improved estimation of carbon stocks and balance in tropical forests. Oecologia 2005, 145, 87–99. [Google Scholar] [CrossRef]
- Kuyah, S.; Dietz, J.; Muthuri, C.; Jamnadass, R.; Mwangi, P.; Coe, R.; Neufeldt, H. Allometric equations for estimating biomass in agricultural landscapes: I. Aboveground biomass. Agric. Ecosyst. Environ. 2012, 158, 216–224. [Google Scholar] [CrossRef]
- Brown, S. Measuring carbon in forests: Current status and future challenges. Environ. Pollut. 2012, 116, 363–372. [Google Scholar] [CrossRef]
- Chave, J.; Condit, R.; Lao, S.; Caspersen, J.P.; Foster, R.B.; Hubbell, S.P. Spatial and temporal variation of biomass in a tropical forest: Results from a large census plot in Panama. J. Ecol. 2003, 91, 240–252. [Google Scholar] [CrossRef]
- Pilli, R.; Anfodillo, T.; Carrer, M. Towards a functional and simplified allometry for estimating forest biomass. For. Ecol. Manag. 2006, 237, 583–593. [Google Scholar] [CrossRef]
- Urquiza-Haas, T.; Dolman, P.M.; Peres, C.A. Regional scale variation in forest structure and biomass in the Yucatan Peninsula, Mexico: Effects of forest disturbance. For. Ecol. Manag. 2007, 247, 80–90. [Google Scholar] [CrossRef]
- Navar, J. Allometric equations for tree species and carbon stocks for forests of northwestern Mexico. For. Ecol. Manag. 2009, 257, 427–434. [Google Scholar] [CrossRef]
- Alvarez, E.; Duque, A.; Saldarriaga, J.; Cabrera, K.; de Las Salas, G.; del Valle, I.; Lema, A.; Moreno, F.; Orrego, S.; Rodriguez, L. Tree above-ground biomass allometries for carbon stocks estimation in the natural forests of Colombia. For. Ecol. Manag. 2012, 267, 297–308. [Google Scholar] [CrossRef]
- Chave, J.; Condit, R.; Aguilar, S.; Hernandez, A.; Lao, S.; Perez, R. Error propagation and scaling for tropical forest biomass estimates. Philos. Trans. R. Soc. B 2004, 359, 409–420. [Google Scholar] [CrossRef]
- Litton, C.M.; Sandquist, D.R.; Cordell, S. Effects of non-native grass invasion on aboveground carbon pools and tree population structure in a tropical dry forest of Hawaii. For. Ecol. Manag. 2006, 231, 105–113. [Google Scholar] [CrossRef]
- Litton, C.M.; Kauffman, J.B. Allometric models for predicting aboveground biomass in two widespread woody plants in Hawaii. Biotropica 2008, 40, 313–320. [Google Scholar] [CrossRef]
- Fonseca, W.; Alice, F.E.; Rey-Benayas, J.M. Carbon accumulation in aboveground and belowground biomass and soil of different age native forest plantations in the humid tropical lowlands of Costa Rica. New For. 2012, 43, 197–211. [Google Scholar]
- Morote, F.A.G.; Serrano, F.R.L.; Andreas, M.; Rubino, E.; Jimenez, J.L.G.; de las Heras, J. Allometries, biomass stocks and biomass allocation in the thermophilic Spanish juniper woodlands of Southern Spain. For. Ecol. Manag. 2012, 270, 85–93. [Google Scholar] [CrossRef]
- Stewart, J.L.; Dunsdon, A.J.; Hellin, J.J.; Hughes, C.E. Wood biomass estimation of Central American dry zone species. In Tropical Forestry Papers; Oxford Forestry Institute: Oxford, UK, 1992; p. 83. [Google Scholar]
- Eamus, D.; McGuinness, K.; Burrows, W. Review of Allometric Relationships for Estimating Woody Biomass for Queensland, the Northern Territory and Western Australia. National Carbon Accounting System (NCAS), Australian Greenhouse Office: Canberra, Australia, 2000. [Google Scholar]
- Openshaw, K. A review of Jatropha curcas: An oil plant of unfulfilled promise. Biomass Bioenerg. 2000, 19, 1–15. [Google Scholar] [CrossRef]
- Rasmussen, L.V.; Rasmussen, K.; Bruun, T.B. Impacts of Jatropha-based biodiesel production on above and belowground carbon stocks. A case study from Mozambique. Energy Policy 2012, 51, 728–736. [Google Scholar] [CrossRef]
- Ghezehei, S.B.; Annandale, J.G.; Everson, C.S. Shoot allometry of Jatropha curcas. South. For. 2009, 71, 279–286. [Google Scholar]
- Verified Carbon Standard—Agriculture, Forestry and Other Land Use Requirements, VCS v3.0 2011. Available online: www.v-c-s.org (accessed on 18 January 2013).
- Achten, W.M.J.; Maes, W.H.; Reubens, B.; Mathijs, E.; Singh, V.P.; Verchot, L.; Muys, B. Biomass production and allocation in Jatropha curcas L. seedlings under different levels of drought stress. Biomass Bioenerg. 2010, 34, 667–676. [Google Scholar] [CrossRef]
- Brassard, B.W.; Chen, H.Y.H.; Bergeron, Y.; Pare, D. Coarse root biomass allometric equations for Abies balsamea, Picea mariana, Pinus banksiana, and Populus tremuloides in the boreal forest of Ontario, Canada. Biomass Bioenerg. 2011, 35, 4189–4196. [Google Scholar] [CrossRef]
- Mponela, P.; Jumbe, C.B.L.; Mwase, W.F. Determinants and extent of land allocation for Jatropha curcas L. cultivation among smallholder farmers in Malawi. Biomass Bioenerg. 2011, 35, 2499–2505. [Google Scholar] [CrossRef]
- Hardcastle, P.D. A Preliminary Silvicultural Classification of Malawi; Forerstry Research Institute of Malawi: Zomba, Malawi, 1978. [Google Scholar]
- West, G.B.; Brown, J.H.; Enquist, B.J. A general model for the structure and allometry of plant vascular systems. Nature 1999, 400, 664–667. [Google Scholar]
- Kaitaniemi, P. Testing allometric scaling laws. J. Theory Biol. 2004, 228, 149–153. [Google Scholar] [CrossRef]
- Djomo, A.N.; Ibrahima, A.; Saborowski, J.; Gravenhorst, G. Allometric equations for biomass estimations in Cameroon and pan moist tropical equations including biomass data from Africa. For. Ecol. Manag. 2010, 260, 1873–1885. [Google Scholar] [CrossRef]
- Parresol, B.R. Assessing tree and stand biomass: A review with examples and critical comparisons. For. Sci. 1999, 45, 573–593. [Google Scholar]
- Ong, J.E.; Gong, W.K.; Wong, C.H. Allometry and partitioning of the mangrove, Rhizophora apiculata. For. Ecol. Manag. 2004, 188, 395–408. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, I.J.; Falster, D.S.; Westoby, M. Bivariate line-fitting methods for allometry. Biol. Rev. 2006, 81, 259–291. [Google Scholar]
- Osborne, J.W. Notes on the use of data transformations. Pract. Assess. Res. Eval. 2002. Available online: http://PAREonline.net/getvn.asp?v=8&n=6 (accessed on 13 March 2013).
- Packard, G.C.; Birchard, G.F.; Boardman, T.J. Fitting statistical models in bivariate allometry. Biol. Rev. 2011, 86, 549–563. [Google Scholar] [CrossRef]
- Cole, G.; Ewel, J.J. Allometric equations for four valuable tropical species. For. Ecol. Manag. 2006, 229, 351–360. [Google Scholar] [CrossRef]
- Ketterings, Q.M.; Coe, R.; Noordwijk, M.; Ambagau, Y.; Palm, C.A. Reducing uncertainty in the use of allometric biomass equations for predicting above-ground tree biomass in mixed secondary forests. For. Ecol. Manag. 2001, 146, 199–209. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Makungwa, S.D.; Chittock, A.; Skole, D.L.; Kanyama-Phiri, G.Y.; Woodhouse, I.H. Allometry for Biomass Estimation in Jatropha Trees Planted as Boundary Hedge in Farmers’ Fields. Forests 2013, 4, 218-233. https://doi.org/10.3390/f4020218
Makungwa SD, Chittock A, Skole DL, Kanyama-Phiri GY, Woodhouse IH. Allometry for Biomass Estimation in Jatropha Trees Planted as Boundary Hedge in Farmers’ Fields. Forests. 2013; 4(2):218-233. https://doi.org/10.3390/f4020218
Chicago/Turabian StyleMakungwa, Stephy D., Abbie Chittock, David L. Skole, George Y. Kanyama-Phiri, and Iain H. Woodhouse. 2013. "Allometry for Biomass Estimation in Jatropha Trees Planted as Boundary Hedge in Farmers’ Fields" Forests 4, no. 2: 218-233. https://doi.org/10.3390/f4020218
APA StyleMakungwa, S. D., Chittock, A., Skole, D. L., Kanyama-Phiri, G. Y., & Woodhouse, I. H. (2013). Allometry for Biomass Estimation in Jatropha Trees Planted as Boundary Hedge in Farmers’ Fields. Forests, 4(2), 218-233. https://doi.org/10.3390/f4020218