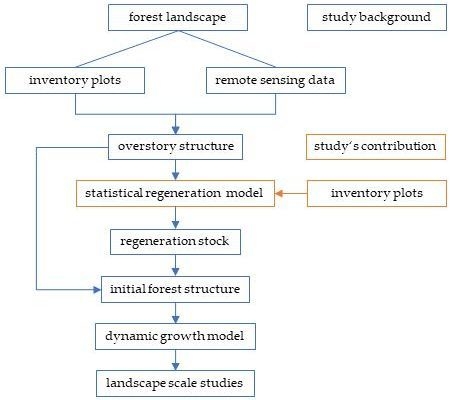
2.1. Data
On the national scale level, the study at hand used the German National Forest Inventory (NFI, 4 km grid) that covers the whole forest area of Germany. The version of these data was the third and most recent survey conducted in 2011 and 2012 [
6] with reference date 1 October 2012 [
21]. For evaluating the bias of the BSFI-based model as related to the resolution of strata, the study applied plot data from the Bavarian State Forest Inventory that covers the forest area managed by the Federal State of Bavaria (BSFI in the further text, 200 m grid). The BSFI surveys 41 forest management units with an average size of about 20,000 ha, each at a repeat cycle of 10 years. Per each forest management unit, it applies individual due dates.
2.1.1. German National Forest Inventory (NFI)
The German National Forest Inventory (description mostly taken from [
6]) is based on a regular 4 km × 4 km quadrangle grid that covers the entire national forest area (~11,400,000 ha, 59,858 plots). It is denser inside some federal states where it has a 2.83 km × 2.83 km or 2 km × 2 km grid. Each grid point represents a north-to-south oriented square with an edge length of 150 m. At the corners of each square, there is exactly one inventory plot, so that the square represents an inventory cluster. On all plots, the NFI measures the overstory (trees of DBH ≥ 7) around each plot center as an angle count sample with a standard basal area factor (BAF) of 4. In addition, it takes the DBH of each sampled tree. Based on the BAF and measured DBH, overstory stem densities per ha are calculated. Within most plots, height measurements are collected on a subset. In continuous cover forests, however, the heights of all sampled trees are measured.
Regeneration trees (DBH < 7 cm) are surveyed within two concentric circles that are situated 5 m north of the plot center. To that end, small saplings with a height of at least 0.2 m and less than 0.5 m are counted on the inner circle of a 1 m radius. Understory trees with a height of at least 0.5 m are counted on the outer circle that has a 2 m radius. Within that circle, the counted trees are attributed to different size classes. The first class comprises trees with a height of less than 1.3 m (and at least 0.5 m). Higher trees are attributed to the DBH classes 0 to 5 cm, 5 to 6 cm, and 6 to 7 cm (upper limits excluded). The radius of the inner circle is extended to 2 m if it would comprise less than four trees within the default radius of 1 m. The NFI registers the browsing of regeneration by an annotation per tree size class. A total of 46% of all plots with a beech-dominated overstory and regeneration had been marked as being browsed by ungulates in at least one regeneration size class.
Within this regeneration survey, the NFI computes regeneration biomass per tree size class from height if the tree considered is less than 1.3 m high and, furthermore, as based on diameter if the tree is higher [
22,
23]. The height-based part of the biomass calculation, to that end, uses one standard height per class, one of 0.35 m (for height class 0.2 to 0.5 m) and one of 0.9 m (>0.5 m to 1.3 m). It applies one generalist biomass to height relation for either coniferous or broadleaved species. On the contrary, the diameter-based biomass estimation interpolates between the height-based biomass at a height of 1.3 m and a species-specific biomass value at a DBH of 10 cm calculated with a modified Marklund model [
24]. For model calibration and evaluation, we selected a subset of 7823 inventory plots with a basal area (BA) share of beech that was more than 55% within the overstory. That data set will be denoted as “NFI data” in the following (
Table 1).
The total BA shares within that set were 84% for beech and 93% for deciduous species in general. We classified 48% of all selected plots as beech monoculture (BA share > 90%). Within the set of beech-dominated plots, the share of regeneration biomass was 74% for beech and 92% for deciduous. Furthermore, 30% of the plots had a regeneration biomass of zero.
2.1.2. Bavarian State Forest Inventory (BSFI)
In contrast to the NFI, which has mainly monitoring purposes, the BSFI is designed to support forest management planning on the level of forest management units (20,000 ha). Therefore, it uses a much higher spatial sampling density on a square grid of a 200 m grid width on average. Each inventory unit is a circular plot with an area of 400 to 500 m
2 that encloses several smaller concentric inventory circles. Only trees above a certain threshold DBH (typically 30 cm) are recorded on the whole plot area. Trees with a DBH < 30 cm and ≥ 11 cm are measured within an 80 to 125 m
2 circle. The smallest class of trees with a DBH < 11 cm, including regeneration trees, is surveyed on a 25 m
2 circle [
25]. The minimum height of regeneration trees to be sampled in the BSFI is 0.2 m, the same as in the NFI. Trees up to a height of 1.3 m are recorded at a higher-class width, i.e., 0.1 m. Trees of a DBH > 0 and < 7 cm are recorded per DBH levels 1.5, 2.5, 3.5, 4.5, 5.5, and 6.5 cm. The beech-dominated BSFI data set used for model calibration and evaluation was selected by the same criteria as the NFI data. That set of plots with beech-dominated overstory will be named “BSFI data” in the following (
Table 2).
The study comprised 11,954 plots from 26 spatially independent inventories over the federal state of Bavaria that had been taken within different years for the period from 2003 to 2012. The basal area shares within the BSFI data were 84% for beech and 91% for deciduous. A total of 41% of all plots were beech monoculture. In order to maintain consistency between NFI and BSFI data, we calculated the regeneration biomass per BSFI plot with the biomass algorithm of the NFI. The share of regeneration biomass then was 76% for beech and 89% for deciduous. Within the BSFI data, 29% of the plots had a regeneration biomass of zero. BSFI plot data, in difference to NFI data, have two topological keys: one that refers to the enclosing forest stand and one that is unique to the enclosing forest management unit. A total of 6073 of the 11,954 plots were spared from model calibration to form strata of a sufficient size for evaluation. Thus, half of all, i.e., 5881 plots remained to parameterize the BSFI-based model.
2.2. Data Preparation
The BSFI surveys regeneration trees within a larger collective of DBH < 11 cm. Again, to keep regeneration tree data from NFI and BSFI consistent with each other, we confined regeneration trees from both inventories to the group with a DBH < 7 cm.
Based on an ecophysiological perception of the regeneration process [
26,
27], we derived a set of overstory characteristics from the inventory plot data that likely determine regeneration biomass. These properties represent above and belowground competition, growth potential, and stand maturity. Both the NFI- and the BSFI-based model use the same set of predictors. In order to parameterize each model, we computed one independent set of plot-related predictor values per each inventory. For quantifying the overstory stand density, we used the Stand Density Index (SDI) [
28] which enables a comparison of stand densities at very different development stages. This structure indicator has been shown to be an effective predictor of regeneration in previous work on experimental plots [
29]. It is defined through Equation (1):
where
N is the number of trees per ha and
Dq is the quadratic mean diameter in cm. The
SDI may be considered to represent the spatial concentration of tree biomass [
4] (p. 399 ff.). Thus, it is a comprehensive indicator of the degree of competition for above- and belowground resources.
While the
SDI expresses the current stand density at the time of measurement, we also required a variable that indicates whether a stand has been kept at higher or lower densities in the long run, because a low density could also result from heavy thinning in a previously dense stand. Therefore, we developed a method for quantifying whether an overstory tree is slender or stout, or has a normal height for its DBH. As a benchmark for normal heights at a given DBH, we fitted an allometric height-diameter model to overstory tree height and diameter (equation with parameters in
Appendix A). Based on the residuals of that model, we estimated a DBH-related probability distribution to standardize the deviation between actual height and normal, i.e., expected height. That distribution enables us to introduce the Height Diameter Characteristic,
HD.
HD is defined as the probability (
P) that any tree of the same diameter might be equally or less high (Equation (2)):
where
h is height as a random variable,
d is DBH as a predictor, and
h0 and
d0 are tree height and DBH as measured, respectively
. Thus,
HD can obtain any value between 0 and 1. For a tree of any diameter,
HD = 0.5 indicates an average, i.e., normal height to diameter ratio.
HD > 0.5 marks an above average tall tree and
HD < 0.5 a stout one. That characteristic, hence, aims to indicate the extent to which a tree had been inhibited in diameter growth through neighbor competition in the past.
The Species Profile Index
SPI [
4] (p. 281 ff.) is a proxy for a forest stand’s richness in species and vertical structure simultaneously. High values indicate structurally rich stands and values of 0 indicate homogenous monospecific stands (Equation (3)):
where
S is the number of species,
Z is the number of height layers, and
p is the relative frequency of species
i in stand height layer
j (exclusively species with
pij > 0 considered). The Species Profile Index in its standard form is based on three stand height layers whose upper borders are
Hmax × 1,
Hmax × 0.8, and
Hmax × 0.5, where
Hmax is the maximum tree height in the stand of interest.
SPI, thus, might indicate whether biomass is concentrated into exactly one tree class or shared among several ones within the stand being considered.
For quantifying the NFI and BSFI plots’ growth potential, we used a Site Index (
SI), defined as the site dependent predominant tree height at age 100. It was computed by the site-productivity algorithm as implemented in the forest growth model SILVA [
16]. That computation was based on the spatial position of the inventory plots and the German map of forest ecological regions [
30]. Moreover, we included the Quadratic Mean Diameter (
Dq) as a predictor that indicates the developmental state of the stand with respect to tree maturity for harvest. Stands with high
Dq are in a phase where regeneration is promoted through felling. German forestry is marked by a large variety of treatment. Within one third of the forest, area transformation to uneven-aged stands and the maintenance of such stands is common. As an indicator of the stand’s development stage, thus, we used the maximum height per plot (
Hmax) that is not limited to even-aged stands. Comparing both inventories by mean and range of corresponding native and derived variables, it can be seen that both are similar (
Table 1 and
Table 2).
2.3. Models
Both models, the NFI- and the BSFI-based version, have the same structure. With the key objective to estimate the local probability distribution of regeneration biomass within an inventory plot, each model comprised a deterministic part as well as a stochastic one. The deterministic module aims to predict the distribution’s expectancy value. As a complement, the stochastic part serves to estimate the variation around that predicted average. At the current stage of development, we aimed to restrict the presumptions for the deterministic module to a minimum. Accordingly, to predict regeneration biomass per ha B, we used a generalized additive mixed model (GAMM) for both the NFI-based model (Equation (4a)) and the BSFI-based one (Equation (4b)):
The fixed effects in this model are the Stand Density Index (
SDI), the Species Profile Index (
SPI), the Height Diameter Characteristic (
HD), the stand maturity indicator (
Hmax), the Site Index (
SI), and the quadratic diameter (
Dq). Except for
Dq, all fixed effects in the model are linear (i.e., they are multiplied by one of the regression parameters
β1,
…,
β5). In contrast, the effect of
Dq is modeled with a nonlinear spline-based smoother [
31], which is indicated by the symbol
s(
Dq). It accounts for a hypothesized nonlinear relation between
Dq and regeneration stock due to a likely minimum of light transmission between the phase of adolescence and intense harvest.
In the model’s NFI-based version, the indices
j and
i associate any plot
j to the corresponding four-plot cluster
i. The BSFI-based version uses a further index
k to represent the embedding of any plot
k into the associated forest stand
j and the enclosing forest management unit
i. Variable
r represents the deterministic part’s residuals, which deserve special attention. Due to the clustered data structure,
r contains random effects on different levels. Within the NFI-based model, it comprises one single group effect that is due to the organization of plots within the inventory clusters (Equation (5)):
where
bi is a cluster specific random effect and
εij represents i.i.d. errors. The BSFI-based model comprises two group effects (Equation (6)):
where
bi is the random effect related to the forest management unit and
bij is the stand-related one.
εijk represents the i.i.d. errors.
For fitting each model’s deterministic part (Equations (4a) resp. (4b)), we took into account that its absolute residuals (
r in Equation (4)) might become larger as regeneration biomass (
B) increases. In order to compensate for such heteroscedasticity, we applied a standard method [
32] (p. 188).
Therefore, the deterministic part of the model (Equations (4a) resp. (4b)) was first calibrated to unweighted data to obtain the corresponding set of absolute residuals, i.e., the values of
r. Then, a specific variance function (Equation (7)) was fitted to the squared residuals:
where
r is the absolute residual,
is the predicted biomass, and
u and
v are parameters to be estimated. Finally, the deterministic model part was fitted again with each observation weighted by the inverse of the residual variance as estimated through Equation (7). For fitting the deterministic part, we used the statistical software R [
33] and the function gamm4 from package gamm4 [
34]. The distribution assumption implied by the fitting procedure for Equation (4a) is
bi ~ N(0,
σ12) and
εij ~ N(0,
σ22). For fitting Equation (4b), we correspondingly assumed
bi ~ N(0,
σ32),
bij ~ N(0,
σ42), and
εijk ~ N(0,
σ52). In order to facilitate the reproduction and application of the deterministic model part, we complemented both Equations (4a) and (4b) with an approximation that refrained from the smoothing spline
s(
Dq). To that end, we replaced
s(
Dq) with a small set of polynomials. Therefore, we predicted regeneration biomass based on the observed
Dq values and the mean of each remainder predictor. Then, we fit one polynomial to the data of predicted biomass over
Dq within each of several
Dq intervals.
The stochastic model part again uses an identical set of equations within the NFI- and the BSFI-based model. It aims to represent the typical spread of regeneration biomass values around an average value as predicted by Equations (4a) resp. (4b). Therefore, it describes the distribution of the deterministic part’s residuals. As these residuals turned out to be heteroscedastic, they were standardized through division by the corresponding predicted mean biomass to obtain relative residuals. For approximating the variability among such relative residuals, the stochastic model part applies the gamma probability density function that is a flexible function confined to positive values. It uses one parameter for shape and one for steepness (rate), as given by Equation (8):
for
R > 0, where
R is the randomly distributed relative residual. Both
α (shape) and
β (rate) are parameters (>0). Г denotes the Gamma function. To obtain parameter values for Equation (8) and to account for remaining heteroscedasticity, we estimated the function’s characteristics from the biomass predicted by the deterministic part. Therefore, the stochastic part uses linear trends of the distribution’s parameters shape
α and rate
β over predicted biomass (Equations (9a) and (9b)):
where
is a biomass value predicted by Equations (4a) resp. (4b);
u1,
u2,
v1, and
v2 are regression parameters; and index
l stands for an observation. Both
ε1 ~ N(
0,
σ62) and
ε2 ~ N(
0,
σ72) are the i.i.d. errors. In order to describe both trends, we divided the range of predicted biomass values into 40 intervals, each representing a quantile width of 2.5%. Within each of them, we fitted one gamma probability density function to the enclosed relative residuals. That way, we obtained a set of 40 distribution functions and, concomitantly, a data set of 40 estimated values for both parameters, shape and rate. For describing the biomass-related trend of a parameter considered—i.e., either shape or rate—we associated each parameter value to the corresponding interval’s predicted biomass median and applied a linear regression (Equations (9a) and (9b)) to the resulting point set. Thus, for any predicted biomass and its accompanying residual distribution, we estimated the corresponding parameters, shape and rate. For fitting the density functions, we used the statistical software R and the function fitdist from package fitdistrplus with moment matching estimation [
35].
2.4. Evaluation
We considered the deterministic model part (Equations (4a) resp. (4b)) as a prototype that might include non-relevant predictors. In order to present each model in a refined form that was exclusively based on relevant fixed effects, we applied two selection criteria to its predictors: one was the significance as obtained from the fitting of Equations (4a) resp. (4b). The other was the Akaike Information Criterion (AIC) [
36]. We used that criterion to identify the model nested into Equations (4a) resp. (4b) that had the minimum tradeoff between goodness of fit and simplicity, i.e., the one with the minimum AIC. Moreover, we assessed the importance of each individual predictor variable using that method (
Appendix B).
For evaluating the stochastic model part, we first tested whether the theoretical density distribution function it uses is a feasible model for the true distribution of the relative residuals obtained from the deterministic module. Although we had expressed each residual of the deterministic part as a relative deviation from the predicted value, we could not eliminate the dependence of the residual distribution on the deterministic part’s predictors. For analyzing the residual distribution, we thus had to use a subrange of the plot data—including observed and predicted biomasses—with an acceptable homogeneity of the residual distribution therein. To that end, we considered a predictor space that was enclosed by a restricted quantile range of the three most influential predictors but still included enough data (quantile 25 to 75%). We then selected all data with predictor values inside that center range, and fitted the gamma density distribution function to the relative residuals obtained from these data. For evaluating that function, we tested it against the subset’s observed relative residuals using the Kolmogorow-Smirnow test of the statistical software R (function ks.test from package stats, [
37]). Moreover, we applied a QQ-plot to plausibilize the function’s fit (R function qqplot from package stats, [
38]).
For evaluating the results generated by the stochastic model part, we simulated relative residuals based on that module. To that end, we predicted the shape and rate of the stochastic part’s gamma distribution function based on each predicted biomass from the deterministic model part (Equations (9a) and (9b)). We then sampled one simulated random residual per biomass value from the resulting set of distribution functions. Finally, we compared the simulated residuals to the observed ones within a QQ-plot.
In order to evaluate the bias of our modelling approach as dependent on the level of stratum differentiation, we applied the BSFI-based model to exemplary strata. We formed these strata from the set of BSFI plots that had been spared from model calibration. On the lower one of two discretization levels considered, the strata exclusively differed in the average tree size as a fundamental stratification criterion. On that level, we formed exactly one stratum per tree size class. Concomitantly, each stratum summarized plots from a number of spatially separate forest management units (each typically 20,000 ha). On the second level of discretization, we further subdivided the strata of the lower discretization level by the forest management unit, to obtain the additional model bias related to such higher spatial differentiation. In order to form the tree size classes, we used a basal area weighted average tree diameter that comprises both understory and overstory. It has routinely been applied to classify strata for SILVA-based simulation scenarios on the landscape scale level (
Table 3). The set of BSFI plots for exemplary stratification was formed through random sampling of at least 140 plots per diameter class and forest management unit.
The study at hand used two evaluation criteria for comparing the modelled distribution of regeneration biomass to the observed one per stratum. One criterion is the mean value. The other is the mean value’s cumulative frequency within the stratum, i.e., the approximate probability of a below average vs. an above-average regeneration biomass. That probability was used to assess whether the stochastic part of the model leads to a realistic frequency of dense vs. sparse regeneration stock within the stratum being considered. For generating the modelled data, we predicted the regeneration biomass per plot based on Equation (4b). Then, we constructed the according residual distribution per plot (Equation (8)) using Equations (9a) and (9b) to estimate the parameters shape and rate in Equation (8) from the plot’s predicted biomass. Finally, we calculated a modelled biomass value per plot as the product of the plot’s predicted biomass and one relative residual value sampled from the estimated distribution. Then, we aggregated the modelled per-plot values on a per-stratum basis to obtain the modelled evaluation criteria. In order to corroborate our per stratum evaluation, we stabilized the value of each evaluation criterion through bootstrapping [
39] with 100 replicates per stratum.