A(H5N1) Virus Evolution in South East Asia
Abstract
:1. Introduction
2. History of emergence and circulation of H5N1 virus in South East Asian countries
2.1. China
2.2. Vietnam
2.3. Thailand
2.4. Cambodia
2.5. Lao PDR
2.6. Myanmar
2.7. Indonesia
2.8. Malaysia
2.9. Comparison of fatality rates in patients from South East Asian countries
| Country | Year of first outbreak | Number of outbreaks in poultry | Number of human infections | Number of human deaths | Country Fatality Rate |
|---|---|---|---|---|---|
| Cambodia | 2003 | 21 | 8 | 7 | 88% |
| China | 1996 | 98 | 38 | 25 | 66% |
| Indonesia | 2003 | 261 | 141 | 115 | 82% |
| Lao PDR | 2003 | 18 | 2 | 2 | 100% |
| Malaysia | 2004 | 16 | 0 | 0 | - |
| Myanmar | 2006 | 93 | 1 | 0 | 0% |
| Thailand | 2004 | 1141 | 25 | 17 | 68% |
| Vietnam | 2003 | 2539 | 111 | 56 | 50% |
3. Evolution of the Predominant Genotypes in South East Asia
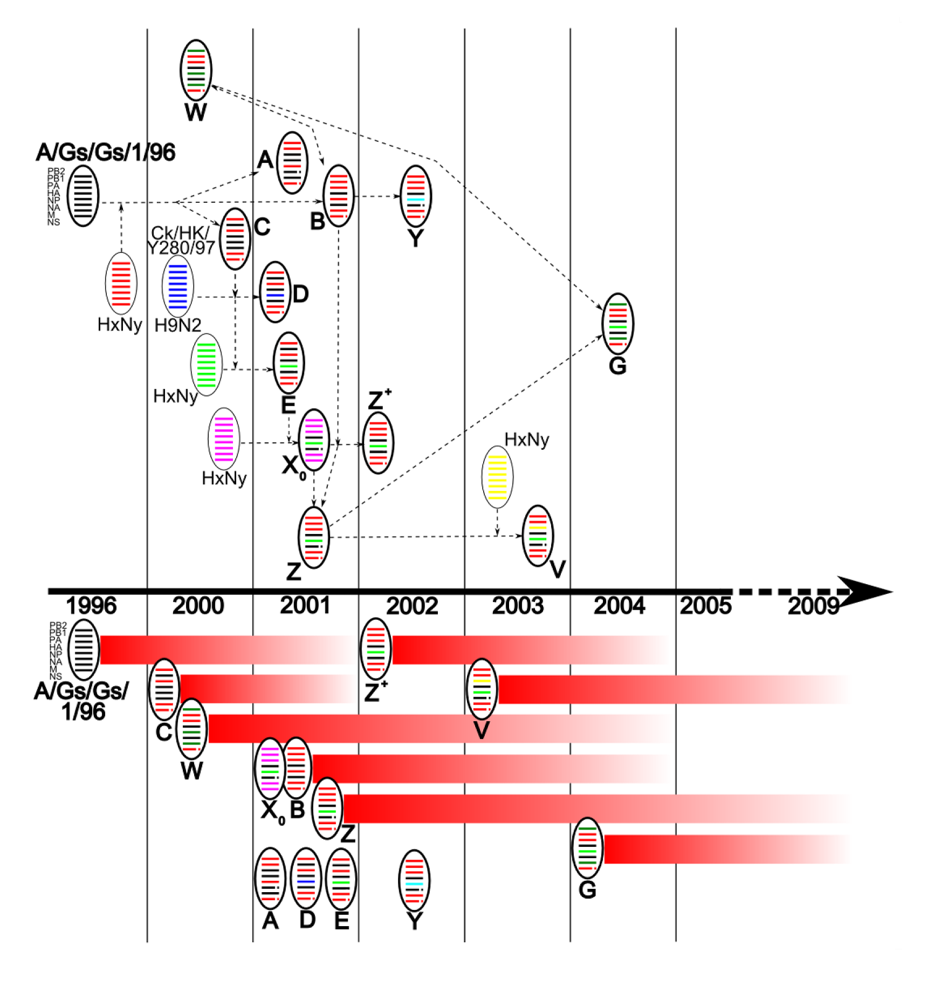

4. H5N1 Clade Evolution in South East Asia

5. Geographical Dynamics of H5N1 Virus Transmission

6. Evolution of Influenza A(H5N1) Virus Genes
6.1. Hemagglutinin (HA) gene
6.2. Neuraminidase (NA) gene
6.3. M gene
6.4. Polymerase complex (PB1, PB2, PA)
6.5. NS1 protein
7. H5N1 virus host range
8. Conclusions
Acknowledgments
References and Notes
- Kruy, S.L.; Buisson, Y.; Buchy, P. Asia: avian influenza H5N1. Bull. Soc. Pathol. Exot. 2008, 101, 238–242. [Google Scholar] [PubMed]
- World Health Organisation. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. 21 04 2009. Available online: http://www.who.int/csr/disease/ avian_influenza/country/cases_table_2009_04_17/en/index.html (accessed 22 April 2009).
- World Health Organisation. H5N1 avian influenza: timeline of major events. 2009. Available online: http://www.who.int/csr/disease/avian_influenza/ai_timeline/en/index.html (accessed 12 June 2009).
- Li, K.S.; Guan, Y.; Wang, J.; Smith, G.J.D.; Xu, K.M.; Duan, L.; Rahardjo, A.P.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; Estoepangestie, A.T.S.; Chaisingh, A.; Auewarakul, P.; Long, H.T.; Hanh, N.T.H.; Webby, R.J.; Poon, L.L.M.; Chen, H.; Shortridge, K.F.; Yuen, K.Y.; Webster, R.G.; Peiris, J.S.M. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 2004, 430, 209–213. [Google Scholar] [CrossRef] [PubMed]
- OIE World Organisation for Animal Health. Avian influenza: H5N1 timeline. 2009. Available online: http://www.oie.int/eng/info_ev/en_AI_factoids_H5N1_Timeline.htm (accessed 12 June 2009).
- Chotpitayasunondh, T.; Ungchusak, K.; Hanshaoworakul, W.; Chunsuthiwat, S.; Sawanpanyalert, P.; Kijphati, R.; Lochindarat, S.; Srisan, P.; Suwan, P.; Osotthanakorn, Y.; Anantasetagoon, T.; Kanjanawasri, S.; Tanupattarachai, S.; Weerakul, J.; Chaiwirattana, R.; Maneerattanaporn, M.; Poolsavathitikool, R.; Chokephaibulkit, K.; Apisarnthanarak, A.; Dowell, S.F. Human disease from influenza A (H5N1), Thailand, 2004. Emerg. Infect. Dis. 2005, 11, 201–209. [Google Scholar] [PubMed]
- Puthavathana, P.; Auewarakul, P.; Charoenying, P.C.; Sangsiriwut, K.; Pooruk, P.; Boonnak, K.; Khanyok, R.; Thawachsupa, P.; Kijphati, R.; Sawanpanyalert, P. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 2005, 86, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.D.; Naipospos, T.S.P.; Nguyen, T.D.; de Jong, M.D.; Vijaykrishna, D.; Usman, T.B.; Hassan, S.S.; Nguyen, T.V.; Dao, T.V.; Bui, N.A.; Leung, Y.H.C.; Cheung, C.L.; Rayner, J.M.; Zhang, J.X.; Zhang, L.J.; Poon, L.L.M.; Li, K.S.; Nguyen, V.C.; Hien, T.T.; Farrar, J.; Webster, R.G.; Chen, H.; Peiris, J.S.M.; Guan, Y. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology 2006, 350, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.J. A review of avian influenza in different bird species. Vet. Microbiol. 2000, 74, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.M.; Rasool, S.T.; Song, D.; Zhu, C.; Hao, Q.; Zhu, Y.; Wu, J. Origin of highly pathogenic H5N1 avian influenza virus in China and genetic characterization of donor and recipient viruses. J. Gen. Virol. 2007, 88, 3094–3099. [Google Scholar] [CrossRef] [PubMed]
- Claas, E.C.; de Jong, J.C.; van Beek, R.; Rimmelzwaan, G.F.; Osterhaus, A.D. Human influenza virus A/HongKong/156/97 (H5N1) infection. Vaccine 1998, 16, 977–978. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, K.; Klimov, A.; Katz, J.; Regnery, H.; Lim, W.; Hall, H.; Perdue, M.; Swayne, D.; Bender, C.; Huang, J.; Hemphill, M.; Rowe, T.; Shaw, M.; Xu, X.; Fukuda, K.; Cox, N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 1998, 279, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Shortridge, K.F.; Stuart-Harris, C.H. An influenza epicentre? Lancet 1982, 2, 812–813. [Google Scholar] [CrossRef]
- Guan, Y.; Peiris, J.S.M.; Lipatov, A.S.; Ellis, T.M.; Dyrting, K.C.; Krauss, S.; Zhang, L.J.; Webster, R.G.; Shortridge, K.F. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 8950–8955. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Guan, Y.; Peiris, M.; Walker, D.; Krauss, S.; Zhou, N.N.; Govorkova, E.A.; Ellis, T.M.; Dyrting, K.C.; Sit, T.; Perez, D.R.; Shortridge, K.F. Characterization of H5N1 influenza viruses that continue to circulate in geese in southeastern China. J. Virol. 2002, 76, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Bahl, J.; Smith, G.; Wang, J.; Vijaykrishna, D.; Zhang, L.; Zhang, J.; Li, K.; Fan, X.; Cheung, C.; Huang, K.; Poon, L.; Shortridge, K.; Webster, R.; Peiris, J.; Chen, H.; Guan, Y. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology 2008, 380, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Webster, R.G. Molecular characterization of H9N2 influenza viruses: were they the donors of the "internal" genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 9363–9367. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Leneva, I.; Krauss, S.; Scholtissek, C.; Chin, P.S.; Peiris, M.; Shortridge, K.F.; Webster, R.G. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J. Virol. 2000, 74, 6309–6315. [Google Scholar] [CrossRef] [PubMed]
- OIE World Organisation for Animal Health. Outbreaks of avian influenza (subtype H5N1) in poultry. From the end of 2003 to 19 April 2009. Available online: http://www.oie.int/downld/AVIAN%20INFLUENZA/Graph%20HPAI/graphs%20HPAI%2019_04_2009.pdf (accessed 12 June 2009).
- Ellis, T.M.; Bousfield, R.B.; Bissett, L.A.; Dyrting, K.C.; Luk, G.S.; Tsim, S.T.; Sturm-Ramirez, K.; Webster, R.G.; Guan, Y.; Malik Peiris, J.S. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 2004, 33, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Sturm-Ramirez, K.M.; Ellis, T.; Bousfield, B.; Bissett, L.; Dyrting, K.; Rehg, J.E.; Poon, L.; Guan, Y.; Peiris, M.; Webster, R.G. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J. Virol. 2004, 78, 4892–4901. [Google Scholar] [CrossRef] [PubMed]
- Bragstad, K.; Jorgensen, P.; Handberg, K.; Hammer, A.; Kabell, S.; Fomsgaard, A. First introduction of highly pathogenic H5N1 avian influenza A viruses in wild and domestic birds in Denmark, northern Europe. Virol. J. 2007, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Olinger, C.M.; Owoade, A.A.; De Landtsheer, S.; Ammerlaan, W.; Niesters, H.G.; Osterhaus, A.D.; Fouchier, R.A.; Muller, C.P. Avian flu: multiple introductions of H5N1 in Nigeria. Nature 2006, 442, 37. [Google Scholar] [CrossRef] [PubMed]
- Molbak, K.; Trykker, H.; Mellergaard, S.; Glismann, S. Avian influenza in Denmark, March-June 2006: public health aspects. Euro. Surveill. 2006, 11, E060615. [Google Scholar] [PubMed]
- Wang, G.; Zhan, D.; Li, L.; Lei, F.; Liu, B.; Liu, D.; Xiao, H.; Feng, Y.; Li, J.; Yang, B.; Yin, Z.; Song, X.; Zhu, X.; Cong, Y.; Pu, J.; Wang, J.; Liu, J.; Gao, G.F.; Zhu, Q. H5N1 avian influenza re-emergence of Lake Qinghai: phylogenetic and antigenic analyses of the newly isolated viruses and roles of migratory birds in virus circulation. J. Gen. Virol. 2008, 89, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vijaykrishna, D.; Duan, L.; Bahl, J.; Zhang, J.X.; Webster, R.G.; Peiris, J.S.M.; Chen, H.; Smith, G.J.D.; Guan, Y. Identification of the progenitors of Indonesian and Vietnamese avian influenza A (H5N1) viruses from southern China. J. Virol. 2008, 82, 3405–3414. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organisation. The regional avian influenza economic assessment workshop Bali, Indonesia, 26 - 29 September 2005 - Vaccination program in Vietnam. 2005. Available online: http://www.fao.org/docrep/009/ag098e/ag098e0a.htm (accessed 12 June 2009). [Google Scholar]
- Wan, X.F.; Nguyen, T.; Davis, C.T.; Smith, C.B.; Zhao, Z.M.; Carrel, M.; Inui, K.; Do, H.T.; Mai, D.T.; Jadhao, S.; Balish, A.; Shu, B.; Luo, F.; Emch, M.; Matsuoka, Y.; Lindstrom, S.E.; Cox, N.J.; Nguyen, C.V.; Klimov, A.; Donis, R.O. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One 2008, 3, e3462. [Google Scholar] [CrossRef] [PubMed]
- Tiensin, T.; Chaitaweesub, P.; Songserm, T.; Chaisingh, A.; Hoonsuwan, W.; Buranathai, C.; Parakamawongsa, T.; Premashthira, S.; Amonsin, A.; Gilbert, M.; Nielen, M.; Stegeman, A. Highly pathogenic avian influenza H5N1, Thailand, 2004. Emerg. Infect. Dis. 2005, 11, 1664–1672. [Google Scholar] [PubMed]
- Buranathai, C.; Amonsin, A.; Chaisigh, A.; Theamboonlers, A.; Pariyothorn, N.; Poovorawan, Y. Surveillance activities and molecular analysis of H5N1 highly pathogenic avian influenza viruses from Thailand, 2004-2005. Avian Dis. 2007, 51, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Seiler, P.; Forrest, H.L.; Khalenkov, A.M.; Franks, J.; Kumar, M.; Karesh, W.B.; Gilbert, M.; Sodnomdarjaa, R.; Douangngeun, B.; Govorkova, E.A.; Webster, R.G. Pathogenicity and vaccine efficacy of different clades of asian H5N1 avian influenza A viruses in domestic ducks. J. Virol. 2008, 82, 11374–11382. [Google Scholar] [CrossRef] [PubMed]
- Tiensin, T.; Nielen, M.; Songserm, T.; Kalpravidh, W.; Chaitaweesub, P.; Amonsin, A.; Chotiprasatintara, S.; Chaisingh, A.; Damrongwatanapokin, S.; Wongkasemjit, S.; Antarasena, C.; Songkitti, V.; Chanachai, K.; Thanapongtham, W.; Stegeman, J.A. Geographic and temporal distribution of highly pathogenic avian influenza A virus (H5N1) in Thailand, 2004-2005: an overview. Avian Dis. 2007, 51, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Chaichoune, K.; Wiriyarat, W.; Thitithanyanont, A.; Phonarknguen, R.; Sariya, L.; Suwanpakdee, S.; Noimor, T.; Chatsurachai, S.; Suriyaphol, P.; Ungchusak, K.; Ratanakorn, P.; Webster, R.G.; Thompson, M.; Auewarakul, P.; Puthavathana, P. Indigenous sources of 2007-2008 H5N1 avian influenza outbreaks in Thailand. J. Gen. Virol. 2009, 90, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Chutinimitkul, S.; Payungporn, S.; Chieochansin, T.; Suwannakarn, K.; Theamboonlers, A.; Poovorawan, Y. The spread of avian influenza H5N1 virus; a pandemic threat to mankind. J. Med. Assoc. Thai. 2006, 89, S218–S233. [Google Scholar] [PubMed]
- Desvaux, S.; Sorn, S.; Holl, D.; Chavernac, D.; Goutard, F.; Thonnat, J.; Porphyre, V.; Menard, C.; Cardinale, E.; Roger, F. HPAI surveillance programme in Cambodia: results and perspectives. Dev. Biol. (Basel) 2006, 124, 211–224. [Google Scholar] [PubMed]
- Ly, S.; Van Kerkhove, M.D.; Holl, D.; Froehlich, Y.; Vong, S. Interaction between humans and poultry, rural Cambodia. Emerg. Infect. Dis. 2007, 13, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Vong, S.; Coghlan, B.; Mardy, S.; Holl, D.; Seng, H.; Ly, S.; Miller, M.J.; Buchy, P.; Froehlich, Y.; Dufourcq, J.B.; Uyeki, T.M.; Lim, W.; Sok, T. Low frequency of poultry-to-human H5NI virus transmission, southern Cambodia, 2005. Emerg. Infect. Dis. 2006, 12, 1542–1547. [Google Scholar] [PubMed]
- Boltz, D.A.; Douangngeun, B.; Sinthasak, S.; Phommachanh, P.; Rolston, S.; Chen, H.; Guan, Y.; Peiris, J.S.; Smith, J.G.; Webster, R.G. H5N1 influenza viruses in Lao People's Democratic Republic. Emerg. Infect. Dis. 2006, 12, 1593–1595. [Google Scholar] [PubMed]
- Kandun, I.N.; Wibisono, H.; Sedyaningsih, E.R. Three Indonesian clusters of H5N1 virus infection in 2005. N. Engl. J. Med. 2006, 355, 2186–2194. [Google Scholar] [CrossRef] [PubMed]
- Butler, D. Family tragedy spotlights flu mutations. Nature 2006, 442, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Tee, K.; Takebe, Y.; Kamarulzaman, A. Emerging and re-emerging viruses in Malaysia, 1997-2007. Int. J. Infect. Dis. 2009, 13, 307–318. [Google Scholar] [CrossRef]
- World Health Organisation. Situation updates - Avian Influenza. Available online: http://www.who.int/csr/disease/avian_influenza/updates/en/index.html (accessed 12 June 2009).
- Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A Virus. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 2008, 358, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Vong, S.; Ly, S.; Van Kerkhove, Maria D.; Achenbach, J.; Holl, D.; Buchy, P.; Sorn, S.; Seng, H.; Uyeki, Timothy M.; Sok, T.; Katz, J.M. Risk factors associated with subclinical human infection with avian influenza A (H5N1) virus - Cambodia, 2006. J. Infect. Dis. 2009, 199, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Vijaykrishna, D.; Bahl, J.; Riley, S.; Duan, L.; Zhang, J.X.; Chen, H.; Peiris, J.S.M.; Smith, G.J.D.; Guan, Y. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 2008, 4, e1000161. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.Y.; Hon, C.C.; Pybus, O.G.; Kosakovsky Pond, S.L.; Wong, R.T.Y.; Yip, C.W.; Zeng, F.; Leung, F.C.C. Evolutionary and transmission dynamics of reassortant H5N1 influenza virus in Indonesia. PLoS Pathog. 2008, 4, e1000130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Smith, G.J.D.; Li, K.S.; Wang, J.; Fan, X.H.; Rayner, J.M.; Vijaykrishna, D.; Zhang, J.X.; Zhang, L.J.; Guo, C.T.; Cheung, C.L.; Xu, K.M.; Duan, L.; Huang, K.; Qin, K.; Leung, Y.H.C.; Wu, W.L.; Lu, H.R.; Chen, Y.; Xia, N.S.; Naipospos, T.S.P.; Yuen, K.Y.; Hassan, S.S.; Bahri, S.; Nguyen, T.D.; Webster, R.G.; Peiris, J.S.M.; Guan, Y. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 2845–2850. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Nguyen, T.; Vijaykrishna, D.; Webster, R.; Guan, Y.; Peiris, J. Multiple sublineages of influenza A virus (H5N1), Vietnam, 2005-2007. Emerg. Infect. Dis. 2008, 14, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Davis, C.T.; Stembridge, W.; Shu, B.; Balish, A.; Inui, K.; Do, H.T.; Ngo, H.T.; Wan, X.F.; McCarron, M.; Lindstrom, S.E.; Cox, N.J.; Nguyen, C.V.; Klimov, A.I.; Donis, R.O. Characterization of a highly pathogenic avian influenza H5N1 virus sublineage in poultry seized at ports of entry into Vietnam. Virology 2009, 387, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Chutinimitkul, S.; Songserm, T.; Amonsin, A.; Payungporn, S.; Suwannakarn, K.; Damrongwatanapokin, S.; Chaisingh, A.; Nuansrichay, B.; Chieochansin, T.; Theamboonlers, A.; Poovorawan, Y. New strain of influenza A virus (H5N1), Thailand. Emerg. Infect. Dis. 2007, 13, 506–507. [Google Scholar] [CrossRef] [PubMed]
- Puthavathana, P.; Sangsiriwut, K.; Korkusol, A.; Pooruk, P.; Auewarakul, P.; Pittayawanganon, C.; Sutdan, D.; Kitphati, R.; Sawanpanyalert, P.; Phommasack, B.; Bounlu, K.; Ungchusak, K. Avian influenza virus (H5N1) in human, Laos. Emerg. Infect. Dis. 2009, 15, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Buchy, P.; Mardy, S.; Vong, S.; Toyoda, T.; Aubin, J.T.; Miller, M.; Touch, S.; Sovann, L.; Dufourcq, J.B.; Richner, B.; Tu, P.V.; Tien, N.T.K.; Lim, W.; Peiris, J.S.M.; Van der Werf, S. Influenza A/H5N1 virus infection in humans in Cambodia. J. Clin. Virol. 2007, 39, 164–168. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Continuing progress towards a unified nomenclature system for the highly pathogenic H5N1 avian influenza viruses. Available online: http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en/ (accessed 12 June 2009).
- Uchida, Y.; Chaichoune, K.; Wiriyarat, W.; Watanabe, C.; Hayashi, T.; Patchimasiri, T.; Nuansrichay, B.; Parchariyanon, S.; Okamatsu, M.; Tsukamoto, K.; Takemae, N.; Ratanakorn, P.; Yamaguchi, S.; Saito, T. Molecular epidemiological analysis of highly pathogenic avian influenza H5N1 subtype isolated from poultry and wild bird in Thailand. Virus Res. 2008, 138, 70–80. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Neighbor-joining (NJ) tree of 1,342 H5N1 HA sequences. 2009. Available online: http://www.who.int/csr/disease/avian_influenza/H5CompleteTree.pdf (accessed 12 June 2009). [Google Scholar]
- Le, M.T.Q.; Wertheim, H.F.L.; Nguyen, H.D.; Taylor, W.; Hoang, P.V.M.; Vuong, C.D.; Nguyen, H.L.K.; Nguyen, H.H.; Nguyen, T.Q.; Nguyen, T.V.; Van, T.D.; Ngoc, B.T.; Bui, T.N.; Nguyen, B.G.; Nguyen, L.T.; Luong, S.T.; Phan, P.H.; Pham, H.V.; Nguyen, T.; Fox, A.; Nguyen, C.V.; Do, H.Q.; Crusat, M.; Farrar, J.; Nguyen, H.T.; de Jong, M.D.; Horby, P. Influenza A H5N1 clade 2.3.4 virus with a different antiviral susceptibility profile replaced clade 1 virus in humans in northern Vietnam. PLoS One 2008, 3, e3339. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, A.M.; Chmura, A.A.; Gibbons, D.W.; Fleischer, R.C.; Marra, P.P.; Daszak, P. Predicting the global spread of H5N1 avian influenza. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 19368–19373. [Google Scholar] [CrossRef] [PubMed]
- Normile, D. Avian influenza. New H5N1 strain emerges in southern China. Science 2006, 314, 742. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Smith, G.J.; Zhang, S.Y.; Qin, K.; Wang, J.; Li, K.S.; Webster, R.G.; Peiris, J.S.; Guan, Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 2005, 436, 191–192. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.D.; Fan, X.H.; Wang, J.; Li, K.S.; Qin, K.; Zhang, J.X.; Vijaykrishna, D.; Cheung, C.L.; Huang, K.; Rayner, J.M.; Peiris, J.S.M.; Chen, H.; Webster, R.G.; Guan, Y. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 16936–16941. [Google Scholar] [CrossRef] [PubMed]
- Amonsin, A.; Choatrakol, C.; Lapkuntod, J.; Tantilertcharoen, R.; Thanawongnuwech, R.; Suradhat, S.; Suwannakarn, K.; Theamboonlers, A.; Poovorawan, Y. Influenza virus (H5N1) in live bird markets and food markets, Thailand. Emerg. Infect. Dis. 2008, 14, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Amonsin, A.; Chutinimitkul, S.; Pariyothorn, N.; Songserm, T.; Damrongwantanapokin, S.; Puranaveja, S.; Jam-On, R.; Sae-Heng, N.; Payungporn, S.; Theamboonlers, A.; Chaisingh, A.; Tantilertcharoen, R.; Suradhat, S.; Thanawongnuwech, R.; Poovorawan, Y. Genetic characterization of influenza A viruses (H5N1) isolated from 3rd wave of Thailand AI outbreaks. Virus Res. 2006, 122, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Buchy, P.; Fourment, M.; Mardy, S.; Sorn, S.; Holl, D.; Ly, S.; Vong, S.; Enouf, V.; Peiris, J.S.M.; van der Werf, S. Molecular Epidemiology of Clade 1 Influenza A Viruses (H5N1), Southern Indochina Peninsula, 2004-2007. Emerg. Infect. Dis. 2009, 15, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Hulse-Post, D.J.; Sturm-Ramirez, K.M.; Humberd, J.; Seiler, P.; Govorkova, E.A.; Krauss, S.; Scholtissek, C.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; Long, H.T.; Naipospos, T.S.P.; Chen, H.; Ellis, T.M.; Guan, Y.; Peiris, J.S.M.; Webster, R.G. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 10682–10687. [Google Scholar] [CrossRef] [PubMed]
- Leung, Y.H.; Zhang, L.J.; Chow, C.K.; Tsang, C.L.; Ng, C.F.; Wong, C.K.; Guan, Y.; Peiris, J.S.M. Poultry drinking water used for avian influenza surveillance. Emerg. Infect. Dis. 2007, 13, 1380–1382. [Google Scholar] [PubMed]
- World Health Organisation. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 2005, 11, 1515–1521. [Google Scholar] [PubMed]
- Takano, R.; Nidom, C.A.; Kiso, M.; Muramoto, Y.; Yamada, S.; Sakai-Tagawa, Y.; Macken, C.; Kawaoka, Y. Phylogenetic characterization of H5N1 avian influenza viruses isolated in Indonesia from 2003-2007. Virology 2009, [Epub, ahead of print]. [Google Scholar]
- Kaverin, N.V.; Rudneva, I.A.; Ilyushina, N.A.; Varich, N.L.; Lipatov, A.S.; Smirnov, Y.A.; Govorkova, E.A.; Gitelman, A.K.; Lvov, D.K.; Webster, R.G. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 2002, 83, 2497–2505. [Google Scholar] [PubMed]
- Wiley, D.C.; Wilson, I.A.; Skehel, J.J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981, 289, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Suzuki, Y.; Suzuki, T.; Le, M.Q.; Nidom, C.A.; Sakai-Tagawa, Y.; Muramoto, Y.; Ito, M.; Kiso, M.; Horimoto, T.; Shinya, K.; Sawada, T.; Kiso, M.; Usui, T.; Murata, T.; Lin, Y.; Hay, A.; Haire, L.F.; Stevens, D.J.; Russell, R.J.; Gamblin, S.J.; Skehel, J.J.; Kawaoka, Y. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 2006, 444, 378. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 11181–11186. [Google Scholar] [CrossRef] [PubMed]
- Shinya, K.; Ebina, M.; Yamada, S.; Ono, M.; Kasai, N.; Kawaoka, Y. Avian flu: influenza virus receptors in the human airway. Nature 2006, 440, 435–436. [Google Scholar] [CrossRef] [PubMed]
- Auewarakul, P.; Chatsurachai, S.; Kongchanagul, A.; Kanrai, P.; Upala, S.; Suriyaphol, P.; Puthavathana, P. Codon volatility of hemagglutinin genes of H5N1 avian influenza viruses from different clades. Virus Genes 2009, 38, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Bender, C.; Hall, H.; Huang, J.; Klimov, A.; Cox, N.; Hay, A.; Gregory, V.; Cameron, K.; Lim, W.; Subbarao, K. Characterization of the surface proteins of influenza A (H5N1) viruses isolated from humans in 1997-1998. Virology 1999, 254, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M.; de Jong, M.D.; Guan, Y. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 2007, 20, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Lackenby, A.; Thompson, C.; Democratis, J. The potential impact of neuraminidase inhibitor resistant influenza. Curr. Opin. Infect. Dis. 2008, 21, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.L.; Herlocher, L.M.; Hoffmann, E.; Matrosovich, M.N.; Monto, A.S.; Webster, R.G.; Govorkova, E.A. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob. Agents Chemother. 2005, 49, 4075–4083. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.D.; Thanh, T.T.; Khanh, T.H.; Hien, V.M.; Smith, G.J.D.; Chau, N.V.; Cam, B.V.; Qui, P.T.; Ha, D.Q.; Guan, Y.; Peiris, J.S.M.; Hien, T.T.; Farrar, J. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 2005, 353, 2667–2672. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.M.; Kiso, M.; Someya, K.; Sakai, Y.T.; Nguyen, T.H.; Nguyen, K.H.L.; Pham, N.D.; Ngyen, H.H.; Yamada, S.; Muramoto, Y.; Horimoto, T.; Takada, A.; Goto, H.; Suzuki, T.; Suzuki, Y.; Kawaoka, Y. Isolation of drug-resistant H5N1 virus. Nature 2005, 437, 1108. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.L.; Ilyushina, N.A.; Salomon, R.; Hoffmann, E.; Webster, R.G.; Govorkova, E.A. Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J. Virol. 2007, 81, 12418–12426. [Google Scholar] [CrossRef] [PubMed]
- McKimm-Breschkin, J.L.; Selleck, P.W.; Bhakti Usman, T.; Johnson, M.A. Reduced sensitivity of influenza A (H5N1) to oseltamivir. Emerg. Infect. Dis. 2007, 13, 1354–1357. [Google Scholar] [PubMed]
- Rameix-Welti, M.A.; Agou, F.; Buchy, P.; Mardy, S.; Aubin, J.T.; Veron, M.; van der Werf, S.; Naffakh, N. Natural variation can significantly alter the sensitivity of influenza A (H5N1) viruses to oseltamivir. Antimicrob. Agents Chemother. 2006, 50, 3809–3815. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.; Bray, M. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008, 78, 91–102. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.D.; Hien, T.T. Avian influenza A (H5N1). J. Clin. Virol. 2006, 35, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Hurt, A.C.; Selleck, P.; Komadina, N.; Shaw, R.; Brown, L.; Barr, I.G. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antiviral Res. 2007, 73, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.L.; Rayner, J.M.; Smith, G.J.D.; Wang, P.; Naipospos, T.S.P.; Zhang, J.; Yuen, K.Y.; Webster, R.G.; Peiris, J.S.M.; Guan, Y.; Chen, H. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J. Infect. Dis. 2006, 193, 1626–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conenello, G.M.; Zamarin, D.; Perrone, L.A.; Tumpey, T.; Palese, P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007, 3, e141. [Google Scholar] [CrossRef]
- Zamarin, D.; Garcia-Sastre, A.; Xiao, X.; Wang, R.; Palese, P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, G.; Dauber, B.; Wolff, T.; Planz, O.; Klenk, H.D.; Stech, J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 18590–18595. [Google Scholar] [CrossRef] [PubMed]
- Shinya, K.; Hamm, S.; Hatta, M.; Ito, H.; Ito, T.; Kawaoka, Y. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 2004, 320, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Steel, J.; Lowen, A.C.; Mubareka, S.; Palese, P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009, 5, e1000252. [Google Scholar] [CrossRef] [PubMed]
- Hulse-Post, D.J.; Franks, J.; Boyd, K.; Salomon, R.; Hoffmann, E.; Yen, H.L.; Webby, R.J.; Walker, D.; Nguyen, T.D.; Webster, R.G. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 2007, 81, 8515–8524. [Google Scholar] [CrossRef] [PubMed]
- Massin, P.; van der Werf, S.; Naffakh, N. Residue 627 of PB2 is a determinant of cold sensitivity in RNA replication of avian influenza viruses. J. Virol. 2001, 75, 5398–5404. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.D.; Simmons, C.P.; Thanh, T.T.; Hien, V.M.; Smith, G.J.D.; Chau, T.N.B.; Hoang, D.M.; Van Vinh Chau, N.; Khanh, T.H.; Dong, V.C.; Qui, P.T.; Van Cam, B.; Ha, D.Q.; Guan, Y.; Peiris, J.S.M.; Chinh, N.T.; Hien, T.T.; Farrar, J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006, 12, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, Y.; Li, Z.; Shi, J.; Shinya, K.; Deng, G.; Qi, Q.; Tian, G.; Fan, S.; Zhao, H.; Sun, Y.; Kawaoka, Y. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 2006, 80, 5976–5983. [Google Scholar] [CrossRef] [PubMed]
- Salomon, R.; Franks, J.; Govorkova, E.A.; Ilyushina, N.A.; Yen, H.L.; Hulse-Post, D.J.; Humberd, J.; Trichet, M.; Rehg, J.E.; Webby, R.J.; Webster, R.G.; Hoffmann, E. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 2006, 203, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Hoffmann, E.; Webster, R.G. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 2002, 8, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.H.; Hoffmann, E.; Webster, R.G. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 2004, 103, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Tian, G.; Li, Y.; Deng, G.; Jiang, Y.; Liu, C.; Liu, W.; Bu, Z.; Kawaoka, Y.; Chen, H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008, 82, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Jiao, P.; Wang, A.; Zhao, F.; Tian, G.; Wang, X.; Yu, K.; Bu, Z.; Chen, H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 2006, 80, 11115–11123. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Yang, H.; Chen, W.; Cao, W.; Zhong, G.; Jiao, P.; Deng, G.; Yu, K.; Yang, C.; Bu, Z.; Kawaoka, Y.; Chen, H. A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J. Virol. 2008, 82, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Obenauer, J.C.; Denson, J.; Mehta, P.K.; Su, X.; Mukatira, S.; Finkelstein, D.B.; Xu, X.; Wang, J.; Ma, J.; Fan, Y.; Rakestraw, K.M.; Webster, R.G.; Hoffmann, E.; Krauss, S.; Zheng, J.; Zhang, Z.; Naeve, C.W. Large-scale sequence analysis of avian influenza isolates. Science 2006, 311, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Ungchusak, K.; Auewarakul, P.; Dowell, S.F.; Kitphati, R.; Auwanit, W.; Puthavathana, P.; Uiprasertkul, M.; Boonnak, K.; Pittayawonganon, C.; Cox, N.J.; Zaki, S.R.; Thawatsupha, P.; Chittaganpitch, M.; Khontong, R.; Simmerman, J.M.; Chunsutthiwat, S. Probable person-to-person transmission of avian influenza A (H5N1). N. Engl. J. Med. 2005, 352, 333–340. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Avian influenza - situation in Indonesia - update 16. Available online: http://www.who.int/csr/don/2006_05_31/en/index.html (accessed 1 September 2009).
- Schultsz, C.; Dong, V.C.; Chau, N.V.; Le, N.T.; Lim, W.; Thanh, T.T.; Dolecek, C.; de Jong, M.D.; Hien, T.T.; Farrar, J. Avian influenza H5N1 and healthcare workers. Emerg. Infect. Dis. 2005, 11, 1158–1159. [Google Scholar] [PubMed]
- Thanh Liem, N.T. Lack of H5N1 avian influenza transmission to hospital employees, Hanoi, 2004. Emerg. Infect. Dis. 2004, 11, 210–215. [Google Scholar]
- Yingst, S.; Saad, M.; Felt, S. Qinghai-like H5N1 from domestic cats, northern Iraq. Emerg. Infect. Dis. 2006, 12, 1295–1297. [Google Scholar] [PubMed]
- Weber, S.; Harder, T.; Starick, E.; Beer, M.; Werner, O.; Hoffmann, B.; Mettenleiter, T.C.; Mundt, E. Molecular analysis of highly pathogenic avian influenza virus of subtype H5N1 isolated from wild birds and mammals in northern Germany. J. Gen. Virol. 2007, 88, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, D. Deadly H5N1 may be brewing in cats. New Scientist 2007, 6–7. [Google Scholar]
- Amonsin, A.; Songserm, T.; Chutinimitkul, S.; Jam-on, R.; Sae-Heng, N.; Pariyothorn, N.; Payungporn, S.; Theamboonlers, A.; Poovorawan, Y. Genetic analysis of influenza A virus (H5N1) derived from domestic cat and dog in Thailand. Arch. Virol. 2007, 152, 1925–1933. [Google Scholar] [CrossRef] [PubMed]
- Keawcharoen, J.; Oraveerakul, K.; Kuiken, T.; Fouchier, R.; Amonsin, A.; Payungporn, S.; Noppornpanth, S.; Wattanodorn, S.; Theamboonlers, A.; Tantilertcharoen, R.; Pattanarangsan, R.; Arya, N.; Ratanakorn, P.; Osterhaus, A.D.M.E.; Poovorawan, Y. Avian influenza H5N1 in tigers and leopards. Emerg. Infect. Dis. 2004, 10, 2189–2191. [Google Scholar] [PubMed]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Gutiérrez, R.A.; Naughtin, M.J.; Horm, S.V.; San, S.; Buchy, P. A(H5N1) Virus Evolution in South East Asia. Viruses 2009, 1, 335-361. https://doi.org/10.3390/v1030335
Gutiérrez RA, Naughtin MJ, Horm SV, San S, Buchy P. A(H5N1) Virus Evolution in South East Asia. Viruses. 2009; 1(3):335-361. https://doi.org/10.3390/v1030335
Chicago/Turabian StyleGutiérrez, Ramona Alikiiteaga, Monica Jane Naughtin, Srey Viseth Horm, Sorn San, and Philippe Buchy. 2009. "A(H5N1) Virus Evolution in South East Asia" Viruses 1, no. 3: 335-361. https://doi.org/10.3390/v1030335
APA StyleGutiérrez, R. A., Naughtin, M. J., Horm, S. V., San, S., & Buchy, P. (2009). A(H5N1) Virus Evolution in South East Asia. Viruses, 1(3), 335-361. https://doi.org/10.3390/v1030335





