Requirement of Cellular Protein CCT7 for the Replication of Fowl Adenovirus Serotype 4 (FAdV-4) in Leghorn Male Hepatocellular Cells Via Interaction with the Viral Hexon Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus and Cells
2.2. Reagents
2.3. Constructs
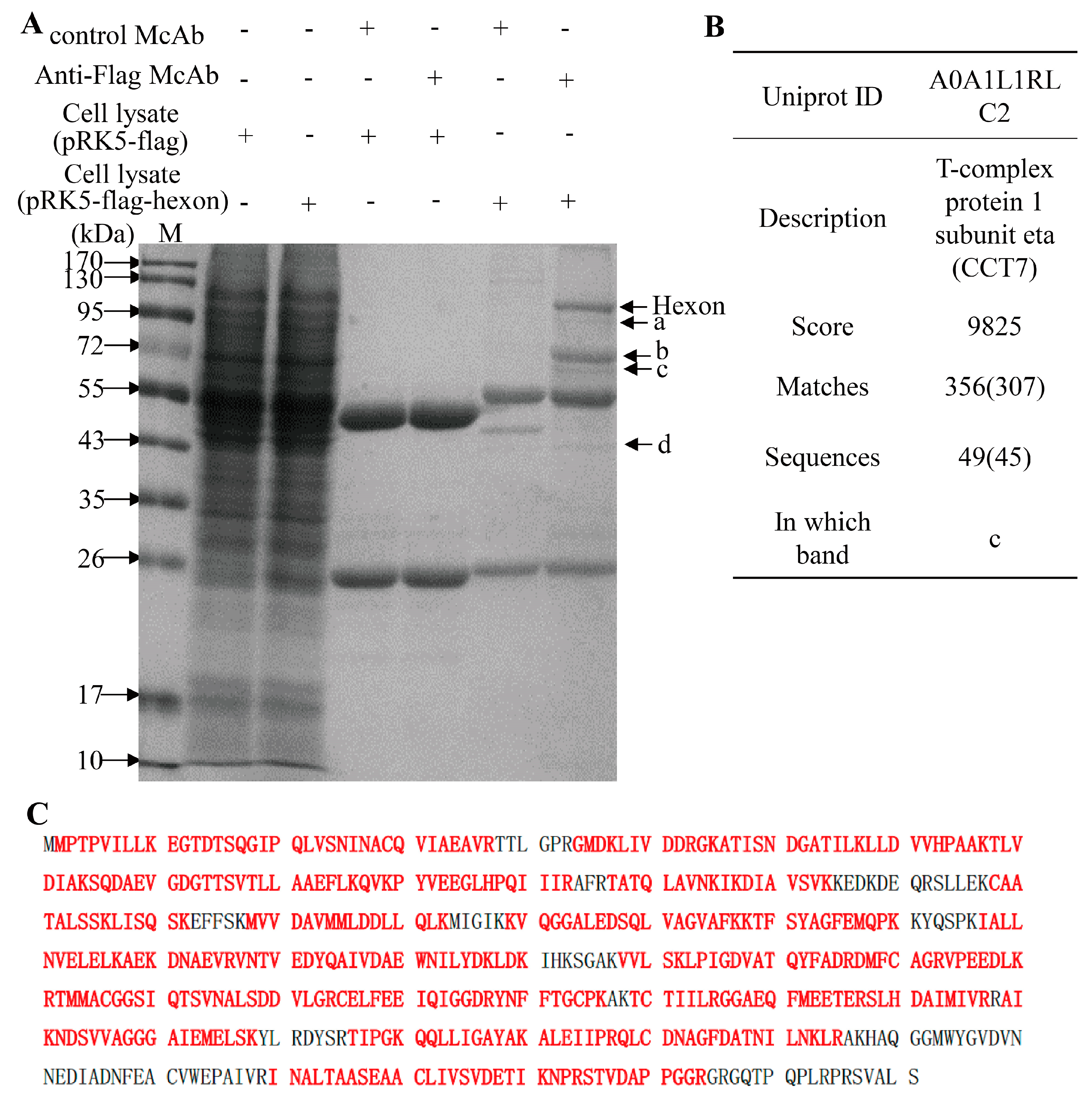
2.4. Pull-Down Assay
2.5. Western Blot Analyses
2.6. Immunoprecipitation
2.7. Transmission Electron Microscopy
2.8. Confocal Laser Scanning Microscopy Assays
2.9. Knockdown of CCT7 by RNAi
2.10. Examination of Hexon in CCT7 Overexpression or Knockdown Cells
2.11. Measurement of FAdV-4 Growth in LMH Cells
2.12. Statistical Analysis
3. Results
3.1. Replication of FAdV-4 HuBWH in LMH cells
3.2. Screening and Identification of FAdV-4 Hexon Interacting Cellular Proteins in LMH Cells
3.3. Hexon Interacts with the Cellular Protein CCT7
3.4. Hexon Colocalizes with CCT7 in Cells
3.5. The Amino Acid Residues 137 to 272 of CCT7 is Responsible for Interaction with the Hexon
3.6. CCT7 is Required for Maintaining the Stability of Hexon in Cells
3.7. CCT7 Facilitates FAdV-4 Replication in Cells
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hess, M. Detection and differentiation of avian adenoviruses: A review. Avian Pathol. 2000, 29, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhong, Q.; Zhao, Y.; Hu, Y.X.; Zhang, G.Z. Pathogenicity and Complete Genome Characterization of Fowl Adenoviruses Isolated from Chickens Associated with Inclusion Body Hepatitis and Hydropericardium Syndrome in China. PLoS ONE 2015, 10, e0133073. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Aziz, T.A.; Al-Attar, M.A. New syndrome in Iraqi chicks. Vet. Rec. 1991, 129, 272. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Nakamura, K.; Tojo, H.; Mase, M.; Shibahara, T.; Yamaguchi, S.; Yuasa, N. Histology, immunohistochemistry, and ultrastructure of hydropericardium syndrome in adult broiler breeders and broiler chicks. Avian Dis. 1998, 42, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Toro, H.; Prusas, C.; Raue, R.; Cerda, L.; Geisse, C.; Gonzalez, C.; Hess, M. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 1999, 43, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.N.; Byun, S.H.; Kim, M.J.; Kim, J.; Sung, H.W.; Mo, I.P. Outbreaks of hydropericardium syndrome and molecular characterization of Korean fowl adenoviral isolates. Avian Dis. 2008, 52, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.J.; Sun, W.; Zhang, G.H.; Qu, Y.J.; Wang, P.F.; Sun, H.L.; Wang, L.; Xiao, Y.H.; Liu, S.D. Hydropericardium syndrome outbreak caused by fowl adenovirus serotype 4 in China in 2015. J. Gen. Virol. 2016, 97, 2684–2690. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Liang, G.; Zhang, J.; Wang, W.; Song, N.; Wang, P.; Zheng, W.; Xie, Q.; Shao, H.; Wan, Z.; et al. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg. Microbes Infect. 2016, 5, e50. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Qiu, L.; Han, Z.; Liu, S. Fowl adenovirus species C serotype 4 is attributed to the emergence of hepatitis-hydropericardium syndrome in chickens in China. Infect. Genet. Evol. 2016, 45, 230–241. [Google Scholar] [CrossRef]
- Shah, M.S.; Ashraf, A.; Khan, M.I.; Rahman, M.; Habib, M.; Chughtai, M.I.; Qureshi, J.A. Fowl adenovirus: History, emergence, biology and development of a vaccine against hydropericardium syndrome. Arch. Virol. 2017, 162, 1833–1843. [Google Scholar] [CrossRef]
- Li, P.H.; Zheng, P.P.; Zhang, T.F.; Wen, G.Y.; Shao, H.B.; Luo, Q.P. Fowl adenovirus serotype 4: Epidemiology, pathogenesis, diagnostic detection, and vaccine strategies. Poult. Sci. 2017, 96, 2630–2640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, R.; Tian, K.; Wang, Z.; Yang, X.; Gao, D.; Zhang, Y.; Fu, J.; Wang, H.; Zhao, J. Fiber2 and hexon genes are closely associated with the virulence of the emerging and highly pathogenic fowl adenovirus 4. Emerg. Microbes Infect. 2018, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Schachner, A.; Marek, A.; Jaskulska, B.; Bilic, I.; Hess, M. Recombinant FAdV-4 fiber-2 protein protects chickens against hepatitis-hydropericardium syndrome (HHS). Vaccine 2014, 32, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yin, L.; Zhou, Q.; Li, Q.; Luo, Y.; Xu, Z.; Zhang, Y.; Xue, C.; Cao, Y. Immunogenicity and protective efficacy of recombinant fiber-2 protein in protecting SPF chickens against fowl adenovirus 4. Vaccine 2018, 36, 1203–1208. [Google Scholar] [CrossRef]
- Bradley, R.R.; Lynch, D.M.; Iampietro, M.J.; Borducchi, E.N.; Barouch, D.H. Adenovirus serotype 5 neutralizing antibodies target both hexon and fiber following vaccination and natural infection. J. Virol. 2012, 86, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Dong, J.; Wang, C.; Zhan, Y.; Zhang, H.; Wu, J.; Kong, W.; Yu, X. Characteristics of neutralizing antibodies to adenovirus capsid proteins in human and animal sera. Virology 2013, 437, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Leitner, A.; Joachimiak, L.A.; Bracher, A.; Monkemeyer, L.; Walzthoeni, T.; Chen, B.; Pechmann, S.; Holmes, S.; Cong, Y.; Ma, B.; et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 2012, 20, 814–825. [Google Scholar] [CrossRef]
- Knee, K.M.; Sergeeva, O.A.; King, J.A. Human TRiC complex purified from HeLa cells contains all eight CCT subunits and is active in vitro. Cell Stress Chaperones 2013, 18, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Llorca, O.; McCormack, E.A.; Hynes, G.; Grantham, J.; Cordell, J.; Carrascosa, J.L.; Willison, K.R.; Fernandez, J.J.; Valpuesta, J.M. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 1999, 402, 693–696. [Google Scholar] [CrossRef]
- Plath, K.; Rapoport, T.A. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J. Cell Biol. 2000, 151, 167–178. [Google Scholar] [CrossRef]
- Camasses, A.; Bogdanova, A.; Shevchenko, A.; Zachariae, W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 2003, 12, 87–100. [Google Scholar] [CrossRef]
- Plimpton, R.L.; Cuellar, J.; Lai, C.W.; Aoba, T.; Makaju, A.; Franklin, S.; Mathis, A.D.; Prince, J.T.; Carrascosa, J.L.; Valpuesta, J.M.; et al. Structures of the Gbeta-CCT and PhLP1-Gbeta-CCT complexes reveal a mechanism for G-protein beta-subunit folding and Gbetagamma dimer assembly. Proc. Natl. Acad. Sci. USA 2015, 112, 2413–2418. [Google Scholar] [CrossRef] [PubMed]
- Melville, M.W.; McClellan, A.J.; Meyer, A.S.; Darveau, A.; Frydman, J. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell Biol. 2003, 23, 3141–3151. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Aizaki, H.; Hara, H.; Matsuda, M.; Ando, T.; Shimoji, T.; Murakami, K.; Masaki, T.; Shoji, I.; Homma, S.; et al. Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology 2011, 410, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Perez, E., Jr.; Xia, A.Y.; Knowlton, J.J.; Cerqueira, F.; Dermody, T.S.; Upton, J.W. Murine cytomegalovirus M72 promotes acute virus replication in vivo and is a substrate of the TRiC/CCT complex. Virology 2018, 522, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Fislova, T.; Thomas, B.; Graef, K.M.; Fodor, E. Association of the influenza virus RNA polymerase subunit PB2 with the host chaperonin CCT. J. Virol. 2010, 84, 8691–8699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, X.; Zan, J.; Wu, Y.; Ye, C.; Ruan, X.; Zhou, J. Cellular chaperonin CCTgamma contributes to rabies virus replication during infection. J. Virol. 2013, 87, 7608–7621. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, J.J.; Fernandez de Castro, I.; Ashbrook, A.W.; Gestaut, D.R.; Zamora, P.F.; Bauer, J.A.; Forrest, J.C.; Frydman, J.; Risco, C.; Dermody, T.S. The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat. Microbiol. 2018, 3, 481–493. [Google Scholar] [CrossRef]
- Niu, Y.; Sun, Q.; Zhang, G.; Liu, X.; Shang, Y.; Xiao, Y.; Liu, S. Fowl adenovirus serotype 4-induced apoptosis, autophagy, and a severe inflammatory response in liver. Vet. Microbiol. 2018, 223, 34–41. [Google Scholar] [CrossRef]
- Li, R.; Li, G.; Lin, J.; Han, S.; Hou, X.; Weng, H.; Guo, M.; Lu, Z.; Li, N.; Shang, Y.; et al. Fowl Adenovirus Serotype 4 SD0828 Infections Causes High Mortality Rate and Cytokine Levels in Specific Pathogen-Free Chickens Compared to Ducks. Front. Immunol. 2018, 9, 49. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Xue, Y.; Li, X.; Cao, H.; Zheng, S.J. Critical role for voltage-dependent anion channel 2 in infectious bursal disease virus-induced apoptosis in host cells via interaction with VP5. J. Virol. 2012, 86, 1328–1338. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Li, X.; Li, X.; Cao, H.; Zheng, S.J. Critical roles of glucocorticoid-induced leucine zipper in infectious bursal disease virus (IBDV)-induced suppression of type I Interferon expression and enhancement of IBDV growth in host cells via interaction with VP4. J. Virol. 2013, 87, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Prusas, C.; Monreal, G. Growth analysis of adenoviruses isolated from pigeons in chicken cells and serological characterization of the isolates. Avian Pathol. 1998, 27, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Yao, K.C.; Liu, Y.Y.; You, G.J.; Li, S.Y.; Liu, P.; Zhao, Q.; Wen Rui Wu, Y.P.; Huang, X.B.; Cao, S.J.; et al. Isolation and molecular characterization of prevalent Fowl adenovirus strains in southwestern China during 2015–2016 for the development of a control strategy. Emerg. Microbes Infect. 2017, 6, e103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zou, Z.; Huang, K.; Lin, X.; Chen, H.; Jin, M. Insights into leghorn male hepatocellular cells response to fowl adenovirus serotype 4 infection by transcriptome analysis. Vet. Microbiol. 2018, 214, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Shibata, T.; Aoshima, M.; Tsubata, T.; Nishida, E. The molecular chaperone TRiC/CCT binds to the Trp-Asp 40 (WD40) repeat protein WDR68 and promotes its folding, protein kinase DYRK1A binding, and nuclear accumulation. J. Biol. Chem. 2014, 289, 33320–33332. [Google Scholar] [CrossRef] [PubMed]
- Kunisawa, J.; Shastri, N. The group II chaperonin TRiC protects proteolytic intermediates from degradation in the MHC class I antigen processing pathway. Mol. Cell 2003, 12, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Sun, Q.; Liu, X.; Liu, S. Mechanism of fowl adenovirus serotype 4-induced heart damage and formation of pericardial effusion. Poult. Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hafirassou, M.L.; Meertens, L.; Umana-Diaz, C.; Labeau, A.; Dejarnac, O.; Bonnet-Madin, L.; Kummerer, B.M.; Delaugerre, C.; Roingeard, P.; Vidalain, P.O.; et al. A Global Interactome Map of the Dengue Virus NS1 Identifies Virus Restriction and Dependency Host Factors. Cell Rep. 2017, 21, 3900–3913. [Google Scholar] [CrossRef]
- Hong, S.; Choi, G.; Park, S.; Chung, A.S.; Hunter, E.; Rhee, S.S. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J. Virol. 2001, 75, 2526–2534. [Google Scholar] [CrossRef]
- Lingappa, J.R.; Martin, R.L.; Wong, M.L.; Ganem, D.; Welch, W.J.; Lingappa, V.R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J. Cell Biol. 1994, 125, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, O.A.; Chen, B.; Haase-Pettingell, C.; Ludtke, S.J.; Chiu, W.; King, J.A. Human CCT4 and CCT5 chaperonin subunits expressed in Escherichia coli form biologically active homo-oligomers. J. Biol. Chem. 2013, 288, 17734–17744. [Google Scholar] [CrossRef] [PubMed]
- Neef, D.W.; Jaeger, A.M.; Gomez-Pastor, R.; Willmund, F.; Frydman, J.; Thiele, D.J. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 2014, 9, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Steinemann, M.; Schlosser, A.; Jank, T.; Aktories, K. The chaperonin TRiC/CCT is essential for the action of bacterial glycosylating protein toxins like Clostridium difficile toxins A and B. Proc. Natl. Acad. Sci. USA 2018, 115, 9580–9585. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Prola, A.; Pires Da Silva, J.; Guilbert, A.; Lecru, L.; Piquereau, J.; Ribeiro, M.; Mateo, P.; Gressette, M.; Fortin, D.; Boursier, C.; et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2alpha deacetylation. Cell Death Differ. 2017, 24, 343–356. [Google Scholar] [CrossRef]
- Bidere, N.; Lorenzo, H.K.; Carmona, S.; Laforge, M.; Harper, F.; Dumont, C.; Senik, A. Cathepsin D triggers Bax activation, resulting in selective apoptosis-inducing factor (AIF) relocation in T lymphocytes entering the early commitment phase to apoptosis. J. Biol. Chem. 2003, 278, 31401–31411. [Google Scholar] [CrossRef]
- Heinrich, M.; Neumeyer, J.; Jakob, M.; Hallas, C.; Tchikov, V.; Winoto-Morbach, S.; Wickel, M.; Schneider-Brachert, W.; Trauzold, A.; Hethke, A.; et al. Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and -3 activation. Cell Death Differ. 2004, 11, 550–563. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, Y.; Hu, X.; Wu, Z.; Guo, W. VPS52 induces apoptosis via cathepsin D in gastric cancer. J. Mol. Med. (Berl) 2017, 95, 1107–1116. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Zhao, M.; Duan, X.; Wang, Y.; Cao, H.; Li, X.; Zheng, S.J. Requirement of Cellular Protein CCT7 for the Replication of Fowl Adenovirus Serotype 4 (FAdV-4) in Leghorn Male Hepatocellular Cells Via Interaction with the Viral Hexon Protein. Viruses 2019, 11, 107. https://doi.org/10.3390/v11020107
Gao J, Zhao M, Duan X, Wang Y, Cao H, Li X, Zheng SJ. Requirement of Cellular Protein CCT7 for the Replication of Fowl Adenovirus Serotype 4 (FAdV-4) in Leghorn Male Hepatocellular Cells Via Interaction with the Viral Hexon Protein. Viruses. 2019; 11(2):107. https://doi.org/10.3390/v11020107
Chicago/Turabian StyleGao, Junfeng, Mingliang Zhao, Xueyan Duan, Yongqiang Wang, Hong Cao, Xiaoqi Li, and Shijun J. Zheng. 2019. "Requirement of Cellular Protein CCT7 for the Replication of Fowl Adenovirus Serotype 4 (FAdV-4) in Leghorn Male Hepatocellular Cells Via Interaction with the Viral Hexon Protein" Viruses 11, no. 2: 107. https://doi.org/10.3390/v11020107
APA StyleGao, J., Zhao, M., Duan, X., Wang, Y., Cao, H., Li, X., & Zheng, S. J. (2019). Requirement of Cellular Protein CCT7 for the Replication of Fowl Adenovirus Serotype 4 (FAdV-4) in Leghorn Male Hepatocellular Cells Via Interaction with the Viral Hexon Protein. Viruses, 11(2), 107. https://doi.org/10.3390/v11020107





