Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Isolation, Purification, and High Titer Phage Stock Preparation
2.3. Host Range Analysis of Isolated Phages
2.4. Amplification of Salmonella Chi-Like Phage Capsid Protein E DNA
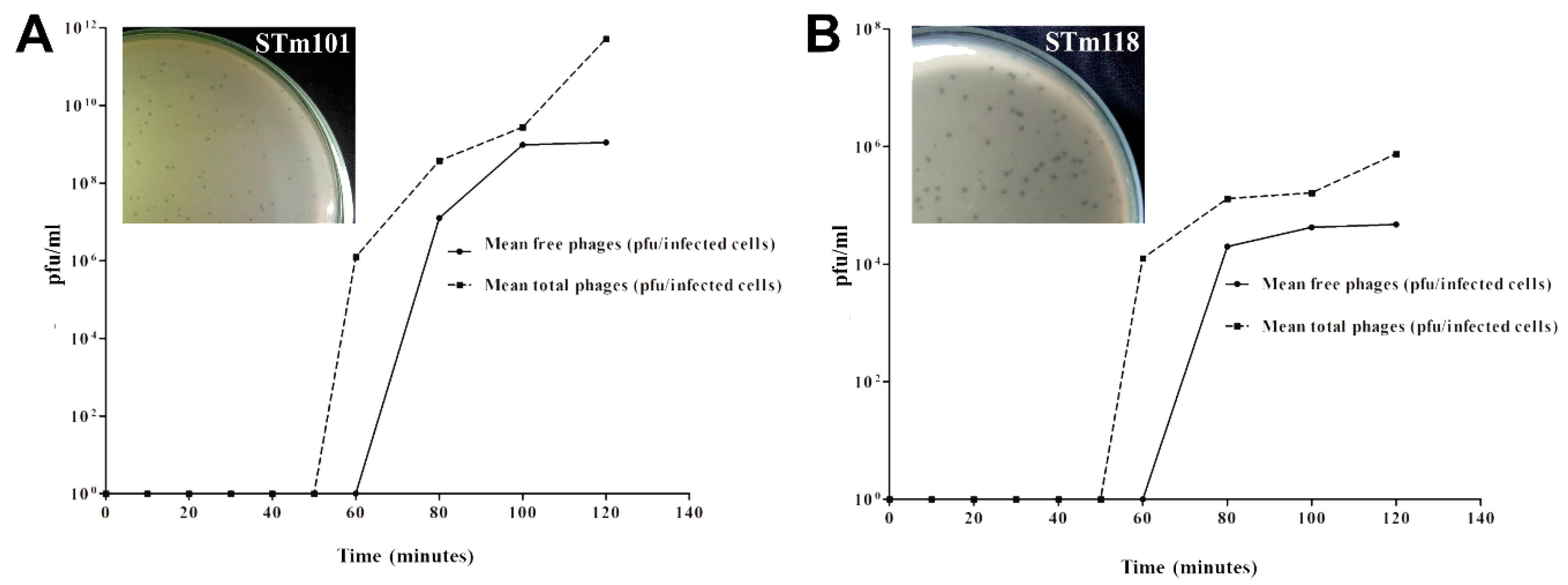
2.5. One-Step Phage Growth Curve Analysis
2.6. Transmission Electron Microscopy
2.7. Next-Generation Sequencing of Phage Genome and Sequences Analysis
2.8. Comparison, Clustering, and Phylogenomic Analysis of Phage Genomes
3. Results
3.1. Host Range Analysis of the Two Thai Phages
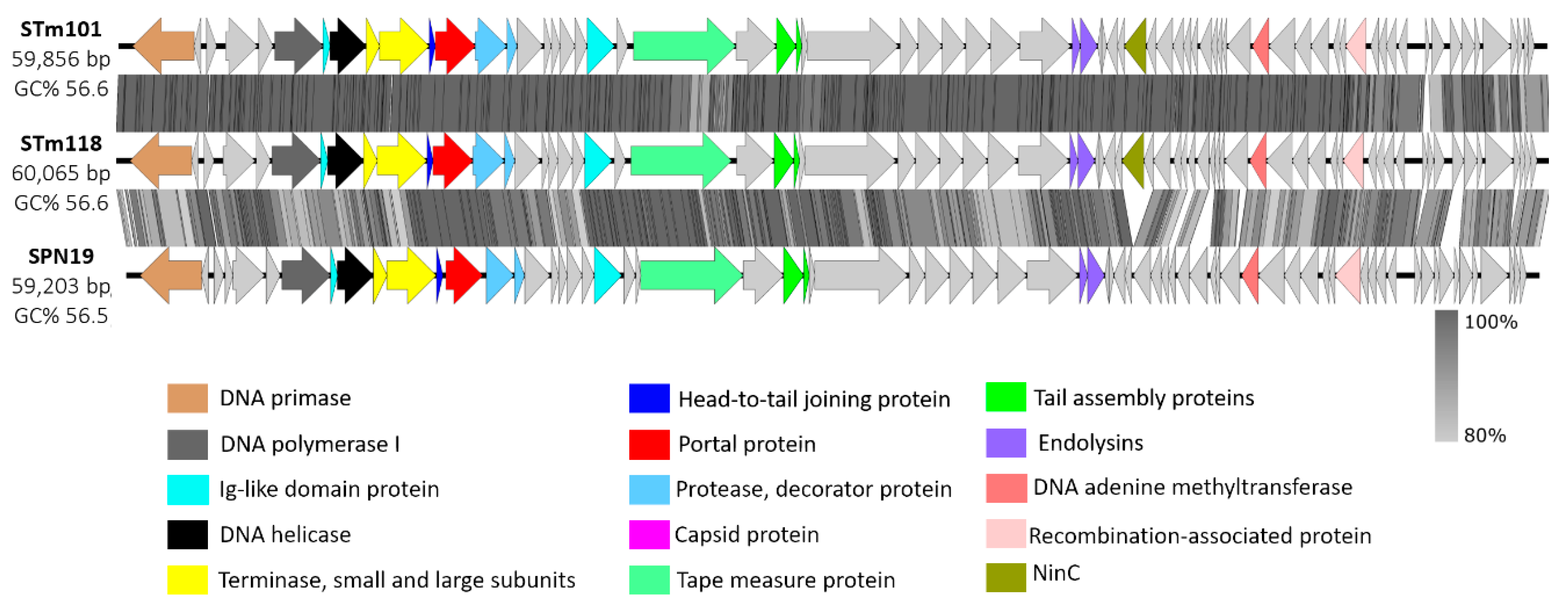
3.2. Genomic Analysis Identifies Thai Phage Isolates as Chi-Like Viruses
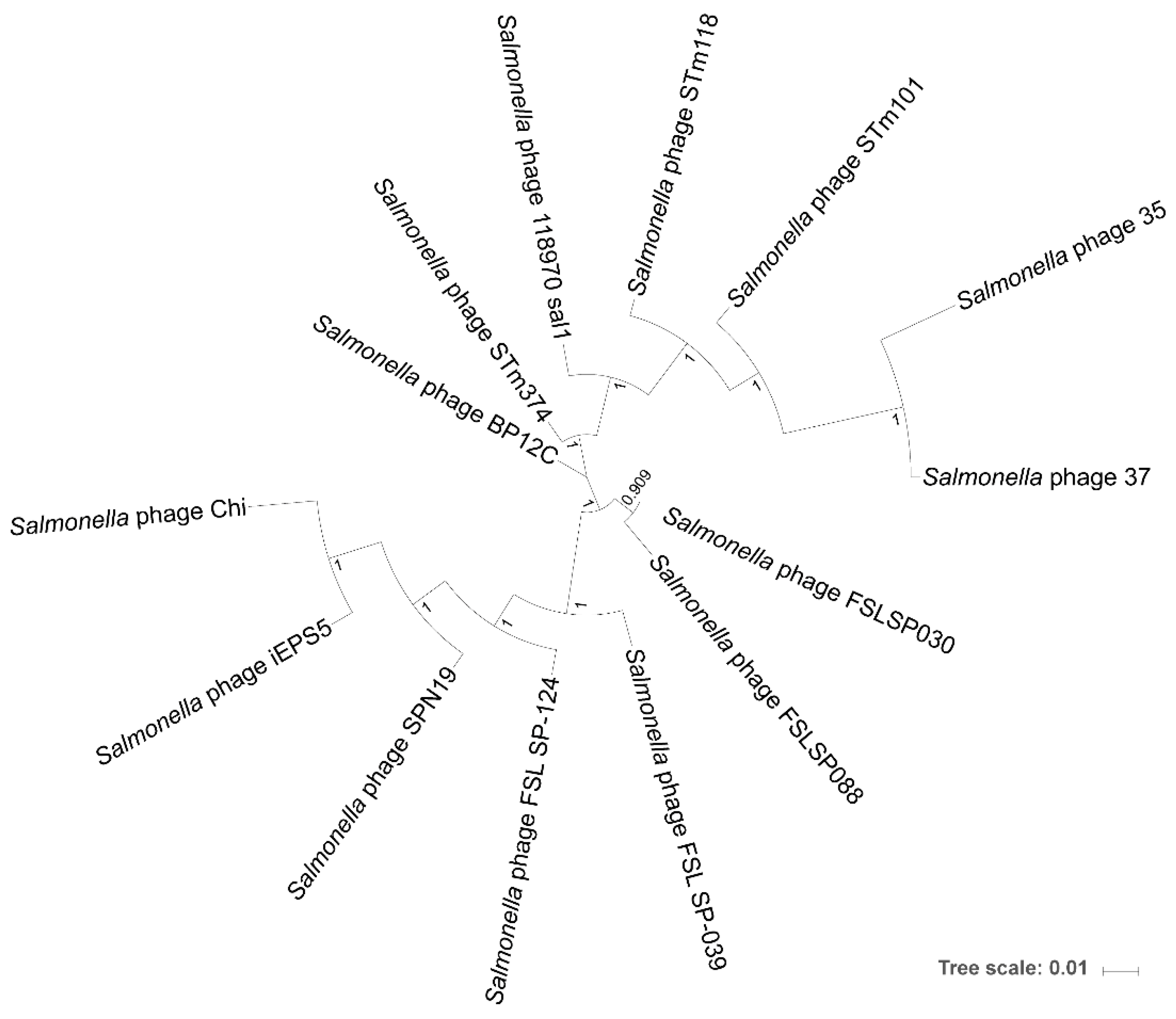
3.3. Phylogenomic Tree Inference of the Chi-Like Phage Cluster
3.4. Chi-Like Salmonella Phages Are Predominant among the Sampled Thai Poultry Farms
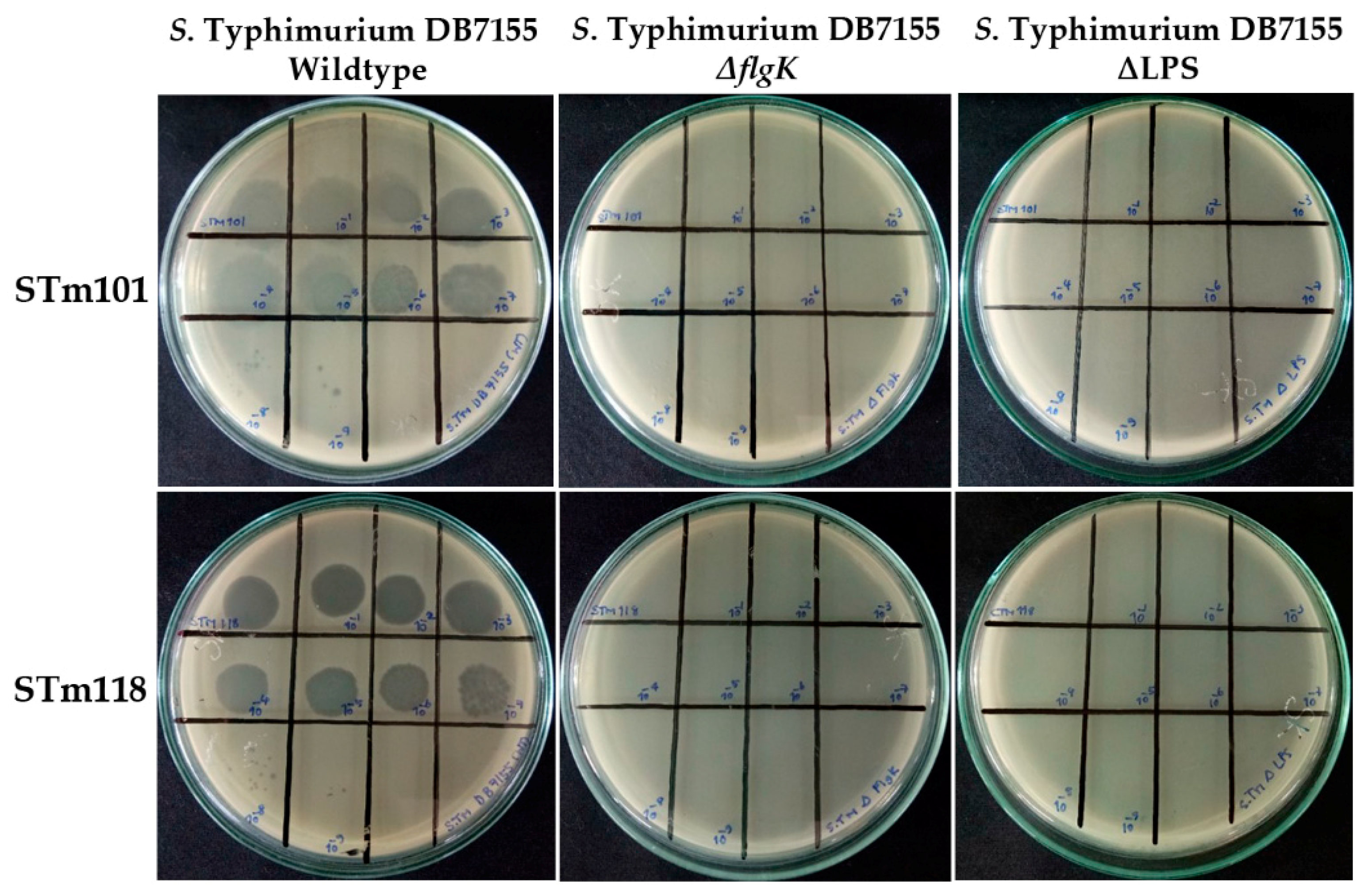
3.5. Flagella and Lipopolysaccharide Are Required for Efficient Infection by the Two Thai Phages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Ethics Approval and Consent to Participate
Conflicts of Interest
Abbreviations
| bp | base pair |
| gp | gene product |
| kbp | kilo-base pair |
| LPS | lipopolysaccharide |
| OM | outer membrane |
| ORFs | open reading frames |
| SM buffer | saline magnesium buffer |
| S. Tm | S. Typhimurium |
| PCR | polymerase chain reaction |
| COG | cluster of orthologous group |
| AAI | average amino acid identity |
| TEM | transmission electron microscopy |
References
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Bruttin, A.; Dillmann, M.L.; Brussow, H. Phage-host interaction: An ecological perspective. J. Bacteriol. 2004, 186, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Glazko, G.; Makarenkov, V.; Liu, J.; Mushegian, A. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct 2007, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Dozois, C.M.; Tinge, S.A.; Lee, T.H.; Curtiss, R., 3rd. Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J. Bacteriol. 2002, 184, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Neal, B.L.; Brown, P.K.; Reeves, P.R. Use of Salmonella phage P22 for transduction in Escherichia coli. J. Bacteriol. 1993, 175, 7115–7118. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.A.; Slauch, J.M. Transduction of low-copy number plasmids by bacteriophage P22. Genetics 1997, 146, 447–456. [Google Scholar] [PubMed]
- Rabsch, W. Salmonella Typhimurium phage typing for pathogens. Methods Mol. Biol. 2007, 394, 177–211. [Google Scholar] [CrossRef] [PubMed]
- Jeoffreys, N.J.; James, G.S.; Chiew, R.; Gilbert, G.L. Practical evaluation of molecular subtyping and phage typing in outbreaks of infection due to Salmonella enterica serotype Typhimurium. Pathology 2001, 33, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Loessner, M.J. Modified bacteriophage tail fiber proteins for labeling, immobilization, capture, and detection of bacteria. Methods Mol. Biol. 2019, 1918, 67–86. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016, 16, 207. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.H.; Horikawa, S.; Bryant, K.; Riggs, R.; Chin, B.A.; Barbaree, J.M. Bacterial assessment of phage magnetoelastic sensors for Salmonella enterica Typhimurium detection in chicken meat. Food Control 2017, 71, 273–278. [Google Scholar] [CrossRef]
- Hungaro, H.M.; Mendonca, R.C.S.; Gouvea, D.M.; Vanetti, M.C.D.; Pinto, C.L.D. Use of bacteriophages to reduce Salmonella in chicken skin in comparison with chemical agents. Food Res. Int. 2013, 52, 75–81. [Google Scholar] [CrossRef]
- Zinno, P.; Devirgiliis, C.; Ercolini, D.; Ongeng, D.; Mauriello, G. Bacteriophage P22 to challenge Salmonella in foods. Int. J. Food Microbiol. 2014, 191, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.D.; Zhang, P.Y.; Zhang, H.; Zhou, Y.; Zhang, L.L.; Wang, R. Bio-control of Salmonella Enteritidis in foods using bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef] [PubMed]
- Waseh, S.; Hanifi-Moghaddam, P.; Coleman, R.; Masotti, M.; Ryan, S.; Foss, M.; MacKenzie, R.; Henry, M.; Szymanski, C.M.; Tanha, J.; et al. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: Prospects of a novel therapy against bacterial infections. PLoS ONE 2010, 5, e13904. [Google Scholar] [CrossRef] [PubMed]
- Sertic, V.; Boulgakov, N.A. Bactériophages spécifiques pour des variétés bactériennes flagellées. C. R. Soc. Biol. 1936, 123, 887–888. [Google Scholar]
- Schmidt, J.M.; Stanier, R.Y. Isolation and characterization of bacteriophages active against stalked bacteria. J. Gen. Microbiol. 1965, 39, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Ferreira, R.C.; Viollier, P.H.; Ely, B.; Poindexter, J.S.; Georgieva, M.; Jensen, G.J.; Wright, E.R. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2011, 108, 9963–9968. [Google Scholar] [CrossRef]
- Meynell, E.W. A phage, phi chi, which attacks motile bacteria. J. Gen. Microbiol. 1961, 25, 253–290. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Ko, C.C.; Jacobs-Sera, D.; Hatfull, G.F.; Erhardt, M.; Hughes, K.T.; Casjens, S.R. Genome sequence of Salmonella phage chi. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Lee, J.H.; Shin, H.; Choi, Y.; Ryu, S. Complete genome sequence analysis of bacterial-flagellum-targeting bacteriophage chi. Arch. Virol. 2013, 158, 2179–2183. [Google Scholar] [CrossRef]
- Switt, A.I.M.; Orsi, R.H.; den Bakker, H.C.; Vongkamjan, K.; Altier, C.; Wiedmann, M. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genom. 2013, 14, 481. [Google Scholar] [CrossRef]
- Kazaks, A.; Dislers, A.; Lipowsky, G.; Nikolajeva, V.; Tars, K. Complete genome sequence of the Enterobacter cancerogenus bacteriophage Enc34. J. Virol. 2012, 86, 11403–11404. [Google Scholar] [CrossRef] [PubMed]
- Onmus-Leone, F.; Hang, J.; Clifford, R.J.; Yang, Y.; Riley, M.C.; Kuschner, R.A.; Waterman, P.E.; Lesho, E.P. Enhanced de novo assembly of high throughput pyrosequencing data using whole genome mapping. PLoS ONE 2013, 8, e61762. [Google Scholar] [CrossRef]
- Joys, T.M. Correlation between susceptibility to bacteriophage PBS1 and motility in Bacillus subtilis. J. Bacteriol. 1965, 90, 1575–1577. [Google Scholar]
- Gonzalez, F.; Helm, R.F.; Broadway, K.M.; Scharf, B.E. More than rotating flagella: lipopolysaccharide as a secondary receptor for flagellotropic phage 7-7-1. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Choi, Y.; Shin, H.; Lee, J.H.; Ryu, S. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 2013, 79, 4829–4837. [Google Scholar] [CrossRef]
- Edwards, S.; Meynell, G.G. The widespred occurrence of enteric flagellar phages. J. Gen. Virol. 1968, 2, 443–444. [Google Scholar] [CrossRef]
- Clokie, M.R.J.; Kropinski, A.M. Bacteriophages: Methods and Protocols, 1st ed.; Humana Press: New York, NY, USA, 2009. [Google Scholar]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Phage classification and characterization. Methods Mol. Biol. 2009, 501, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, R.; Liu, C.M.; Leung, C.M.; Ting, H.F.; Sadakane, K.; Yamashita, H.; Lam, T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Coordinators, N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017, 45. [Google Scholar] [CrossRef]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. (2013). Available online: https://www.R-project.org/ (accessed on 17 May 2017).
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Shimodaira, H.; Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef]
- McLaughlin, M.R.; King, R.A. Characterization of Salmonella bacteriophages isolated from swine lagoon effluent. Curr. Microbiol. 2008, 56, 208–213. [Google Scholar] [CrossRef]
- Schade, S.Z.; Adler, J.; Ris, H. How bacteriophage chi attacks motile bacteria. J. Virol. 1967, 1, 599–609. [Google Scholar]
- Paradiso, R.; Lombardi, S.; Iodice, M.G.; Riccardi, M.G.; Orsini, M.; Bolletti Censi, S.; Galiero, G.; Borriello, G. Complete genome sequences of three Siphoviridae bacteriophages infecting Salmonella enterica serovar Enteritidis. Genome Announc. 2016, 4, e00939-16. [Google Scholar] [CrossRef]
- Karpe, Y.A.; Kanade, G.D.; Pingale, K.D.; Arankalle, V.A.; Banerjee, K. Genomic characterization of Salmonella bacteriophages isolated from India. Virus Genes 2016, 52, 117–126. [Google Scholar] [CrossRef]
- Jiang, S.C.; Paul, J.H. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 1998, 64, 2780–2787. [Google Scholar]
- Paul, J.H. Microbial gene transfer: an ecological perspective. J. Mol. Microbiol. Biotechnol. 1999, 1, 45–50. [Google Scholar]
- Rohwer, F.; Edwards, R. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 2002, 184, 4529–4535. [Google Scholar] [CrossRef]
- Lindell, D.; Sullivan, M.B.; Johnson, Z.I.; Tolonen, A.C.; Rohwer, F.; Chisholm, S.W. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 11013–11018. [Google Scholar] [CrossRef]
- Grose, J.H.; Casjens, S.R. Understanding the enormous diversity of bacteriophages: The tailed phages that infect the bacterial family Enterobacteriaceae. Virology 2014, 468–470, 421–443. [Google Scholar] [CrossRef]
- Bender, R.A.; Refson, C.M.; O’Neill, E.A. Role of the flagellum in cell-cycle-dependent expression of bacteriophage receptor activity in Caulobacter crescentus. J. Bacteriol. 1989, 171, 1035–1040. [Google Scholar] [CrossRef]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol. Microbiol. 2013, 87, 818–834. [Google Scholar] [CrossRef]
- Andres, D.; Baxa, U.; Hanke, C.; Seckler, R.; Barbirz, S. Carbohydrate binding of Salmonella phage P22 tailspike protein and its role during host cell infection. Biochem. Soc. Trans. 2010, 38, 1386–1389. [Google Scholar] [CrossRef]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef]
- Washizaki, A.; Yonesaki, T.; Otsuka, Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiologyopen 2016, 5, 1003–1015. [Google Scholar] [CrossRef]
- Trojet, S.N.; Caumont-Sarcos, A.; Perrody, E.; Comeau, A.M.; Krisch, H.M. The gp38 adhesins of the T4 superfamily: A complex modular determinant of the phage’s host specificity. Genome Biol. Evol. 2011, 3, 674–686. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Loessner, M.J.; Hagens, S.; Slijkhuis, A.J.H.; Klumpp, J.A.; Marti, R. Bacteriophage for biocontrol of Salmonella and in the manufacturing or processing of foods. Patent Application No. 14/399,383, 7 May 2015. [Google Scholar]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; Burd, I.C.E.D.; et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Nilsson, A.S. Phage therapy-constraints and possibilities. Ups. J. Med. Sci. 2014, 119, 192–198. [Google Scholar] [CrossRef]
- Kilcher, S.; Loessner, M.J. Engineering bacteriophages as versatile biologics. Trends Microbiol. 2019, 27, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically engineered phages: A review of advances over the last decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Mavrich, T.N.; Hatfull, G.F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2017, 2, 17112. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Arisaka, F.; van Raaij, M.J.; Kostyuchenko, V.A.; Aksyuk, A.A.; Kanamaru, S.; Rossmann, M.G. Morphogenesis of the T4 tail and tail fibers. Virol. J. 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Denyes, J.M.; Arndt, H.; Loessner, M.J.; Leiman, P.G.; Klumpp, J. Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition. Structure 2018, 26, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Thomassen, E.; Gielen, G.; Schutz, M.; Schoehn, G.; Abrahams, J.P.; Miller, S.; van Raaij, M.J. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 2003, 331, 361–373. [Google Scholar] [CrossRef]

| Salmonella Serovar (s) | No. of Isolates | Spot Lysis (% Infectivity) of Phages | |
|---|---|---|---|
| STm101 | STm118 | ||
| S. Enteritidis | 32 | 3 (9.4%) | 21 (65.6%) |
| S. Typhimurium | 27 | 20 (74.1%) | 17 (63%) |
| S. Virchow | 15 | 7 (46.7%) | 4 (26.7%) |
| S. Hadar | 11 | 3 (27.3%) | 2 (18.2%) |
| S. Choleraesuis | 13 | 10 (76.9%) | 6 (46.2%) |
| S. Agona | 3 | 3 (100%) | 3 (100%) |
| S. Saintpaul | 1 | 1 (100%) | 1 (100%) |
| S. Kentucky | 1 | 0 (0%) | 1 (100%) |
| S. Albany | 3 | 0 (0%) | 0 (0%) |
| S. I 4,[5],12:i:- | 4 | 0 (0%) | 0 (0%) |
| S. Altona | 1 | 0 (0%) | 0 (0%) |
| S. Corvallis | 2 | 0 (0%) | 0 (0%) |
| S. Give | 3 | 0 (0%) | 0 (0%) |
| S. Schwarzengrund | 1 | 0 (0%) | 0 (0%) |
| S. Singapore | 1 | 0 (0%) | 0 (0%) |
| Total | 118 | 47 (39.8%) | 55 (46.6%) |
| Control strains | |||
| Listeria monocytogenes | 13 | 0 (0%) | 0 (0%) |
| Vibrio spp. | 8 | 0 (0%) | 0 (0%) |
| Escherichia coli | 5 | 0 (0%) | 0 (0%) |
| Staphylococcus aureus | 5 | 0 (0%) | 0 (0%) |
| Total | 31 | 0 (0%) | 0 (0%) |
| Phage | Accession no. | Size (bp) | GC (%) | CDSs | Avg CDS Size (bp) | tRNAs | Avg COG Size | Family | Bacterial Host | Source | Country | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella phage STm101 | KX765862 | 59,856 | 56.6 | 71 | 716 | 0 | 16.8 | Siphoviridae | S. Typhimurium | Chicken feces | Thailand | This study |
| Salmonella phage STm118 | KX765863 | 60,065 | 56.6 | 72 | 624 | 0 | 16.6 | Siphoviridae | S. Typhimurium | Chicken feces | Thailand | This study |
| Salmonella phage STm374 | KX765864 | 59,934 | 56.6 | 72 | 502 | 0 | 16.2 | Siphoviridae | S. Typhimurium | Chicken feces | Thailand | - |
| Salmonella phage 118970_sal1 | NC_031930.1 | 59,518 | 56.5 | 71 | 782 | 0 | 11.5 | Siphoviridae | S. Enteritidis | Water buffalo feces | Italy | [48] |
| Salmonella phage 35 | KR296689.1 | 55,391 | 56.8 | 91 | 561 | 0 | 7 | Podoviridae | S. Gallinarum | Sewage | India | [49] |
| Salmonella phage 37 | NC_029045.1 | 60,216 | 56.5 | 105 | 520 | 0 | 6.3 | Siphoviridae | S. Gallinarum | Sewage | India | [49] |
| Salmonella phage BP12C | NC_031228.1 | 60,606 | 56.4 | 76 | 757 | 0 | 11.7 | Siphoviridae | S. Hadar | Sewage | USA | - |
| Salmonella phage Chi | JX094499.1; KM458633.1 | 59,407 | 56.5 | 75 | 749 | 0 | 11.9 | Siphoviridae | Salmonella enterica, E. coli | - | France | [21,22] |
| Salmonella phage FSL SP-039 | KC139514 | 59,815 | 56.6 | 71 | 776 | 0 | 12.3 | Siphoviridae | S. Cerro | Dairy farms | USA | [23] |
| Salmonella phage FSL SP-124 | KC139515 | 59,245 | 56.5 | 71 | 765 | 0 | 12.2 | Siphoviridae | S. Cerro | Dairy farms | USA | [23] |
| Salmonella phage FSL SP-030 | NC_021779.1 | 59,746 | 56.6 | 71 | 776 | 0 | 12.3 | Siphoviridae | S. Dublin | Dairy farms | USA | [23] |
| Salmonella phage FSL SP-088 | NC_021780.1 | 59,454 | 56.4 | 70 | 779 | 0 | 12.3 | Siphoviridae | S. Typhimurium | Dairy farms | USA | [23] |
| Salmonella phage iEPS5 | NC_021783.1 | 59,254 | 56.3 | 73 | 765 | 0 | 12 | Siphoviridae | S. Typhimurium | Sewage | South Korea | [28] |
| Salmonella phage SPN19 | NC_019417.1 | 59,203 | 56.5 | 72 | 770 | 0 | 12.1 | Siphoviridae | S. Typhimurium | Sewage | South Korea | - |
| S. Typhimurium | Other Designations and Features | Source | STm101 | STm118 |
|---|---|---|---|---|
| Wildtype | DB7155 | [56] | ++ | ++ |
| ΔompA | DB7155, ΔompA::Kanr; outer membrane protein A | [56] | ++ | ++ |
| ΔompC | DB7155, ΔompC::Kanr; outer membrane protein C | [56] | ++ | ++ |
| ΔompX | DB7155, ΔompX::Kanr; outer membrane protein X | [56] | ++ | ++ |
| ΔbtuB | DB7155, ΔbtuB::Kanr; TonB-dependent vitamin B12 transporter | [56] | ++ | ++ |
| ΔfadL | DB7155, ΔfadL::Kanr; long-chain fatty acids transporter | [56] | ++ | ++ |
| Δtsx | DB7155, Δtsx::Kanr; nucleoside-specific outer membrane channel | [56] | ++ | ++ |
| ΔtonB | DB7155, ΔtonB::Kanr; active transport regulator protein TonB | [56] | ++ | ++ |
| ΔflgK | SL1344_CH502, ΔflgK::Kanr; hook-filament junction protein | [28] | - | - |
| ΔLPS | DB7155 ΔLPS::Cmr; all LPS synthesis genes for regions distal to KDO residues deleted. | [56] | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phothaworn, P.; Dunne, M.; Supokaivanich, R.; Ong, C.; Lim, J.; Taharnklaew, R.; Vesaratchavest, M.; Khumthong, R.; Pringsulaka, O.; Ajawatanawong, P.; et al. Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms. Viruses 2019, 11, 520. https://doi.org/10.3390/v11060520
Phothaworn P, Dunne M, Supokaivanich R, Ong C, Lim J, Taharnklaew R, Vesaratchavest M, Khumthong R, Pringsulaka O, Ajawatanawong P, et al. Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms. Viruses. 2019; 11(6):520. https://doi.org/10.3390/v11060520
Chicago/Turabian StylePhothaworn, Preeda, Matthew Dunne, Rattaya Supokaivanich, Catherine Ong, Jiali Lim, Rutjawate Taharnklaew, Mongkol Vesaratchavest, Rabuesak Khumthong, Onanong Pringsulaka, Pravech Ajawatanawong, and et al. 2019. "Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms" Viruses 11, no. 6: 520. https://doi.org/10.3390/v11060520
APA StylePhothaworn, P., Dunne, M., Supokaivanich, R., Ong, C., Lim, J., Taharnklaew, R., Vesaratchavest, M., Khumthong, R., Pringsulaka, O., Ajawatanawong, P., Klumpp, J., Brown, N., Imam, M., Clokie, M. R. J., Galyov, E. E., & Korbsrisate, S. (2019). Characterization of Flagellotropic, Chi-Like Salmonella Phages Isolated from Thai Poultry Farms. Viruses, 11(6), 520. https://doi.org/10.3390/v11060520





