HIV-1 Mutant Assembly, Processing and Infectivity Expresses Pol Independent of Gag
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Cell Culture, Transfection, and Infection
2.3. Sucrose Density Gradient Fractionation
2.4. Western Immunoblot Analysis
3. Results
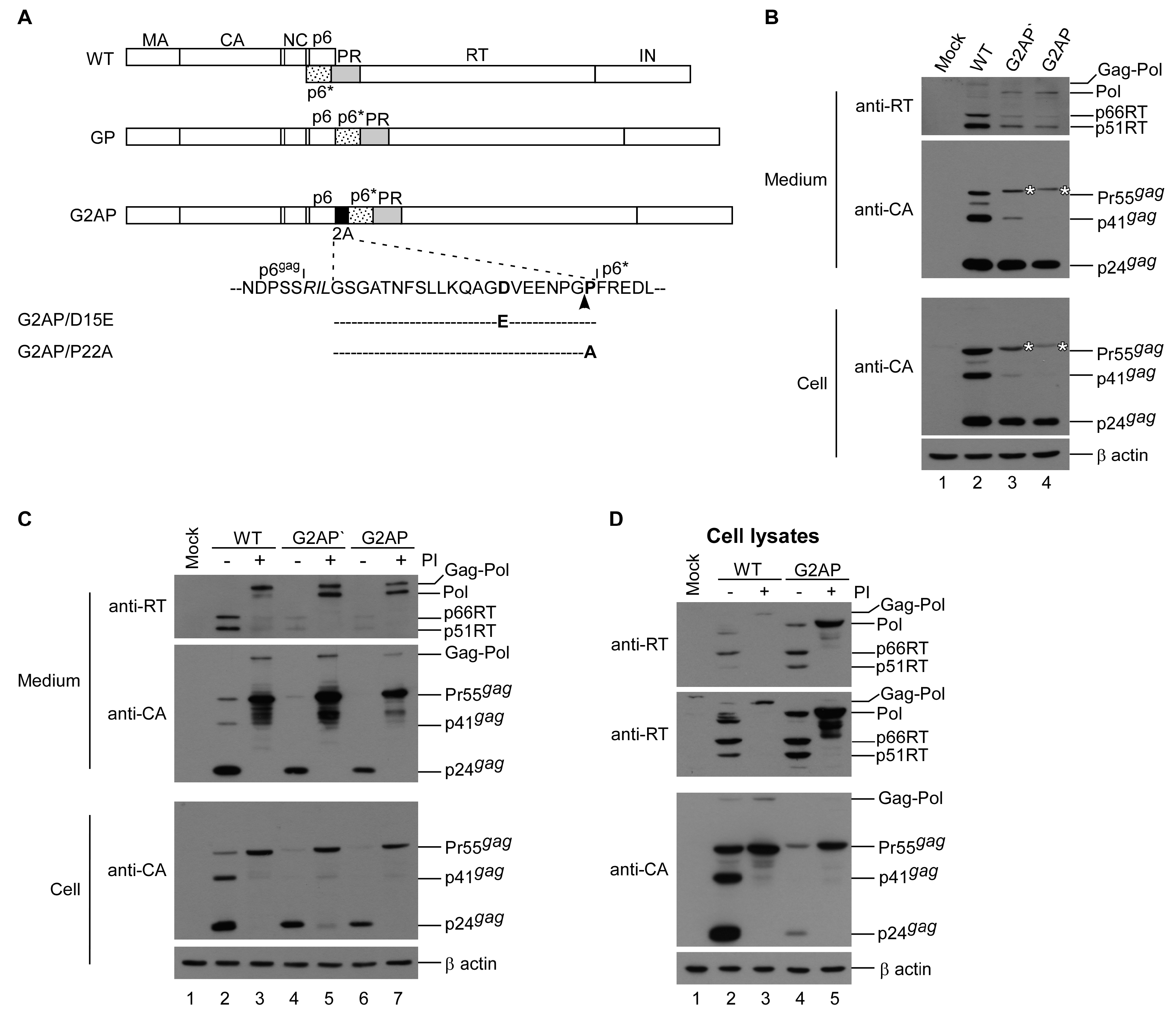
3.1. HIV-1 Gag and Pol Expression in a Single Plasmid
3.2. G2AP Exhibits Wild-Type HIV-1 Particle Density and Possesses Infectivity
3.3. Reduced Pol Expression Significantly Increases Virus Yields
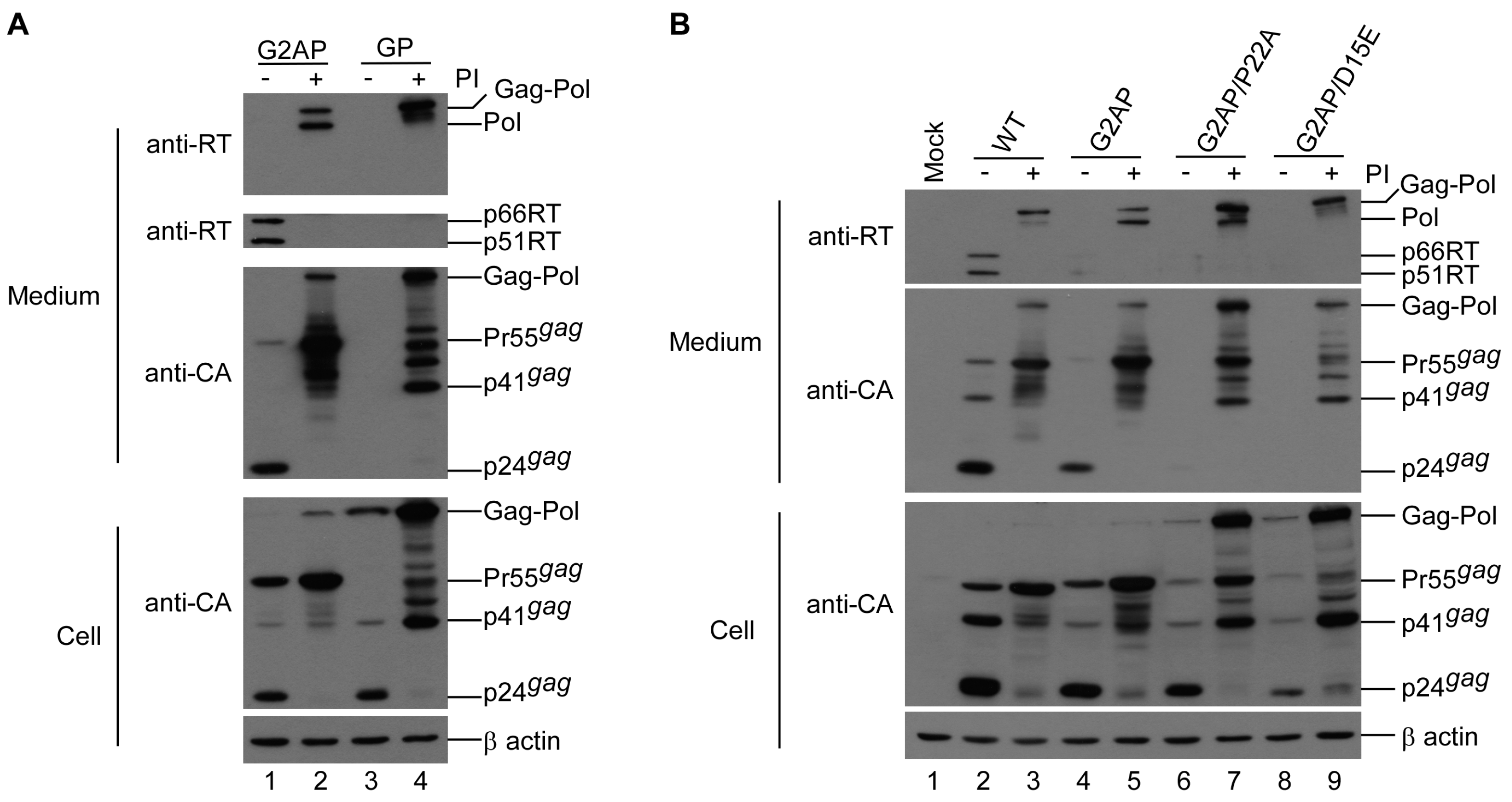
3.4. PR Activity Attenuation Increases Virus Titers
3.5. PR-RT Cleavage Blocking Enhances G2AP Virus Yields
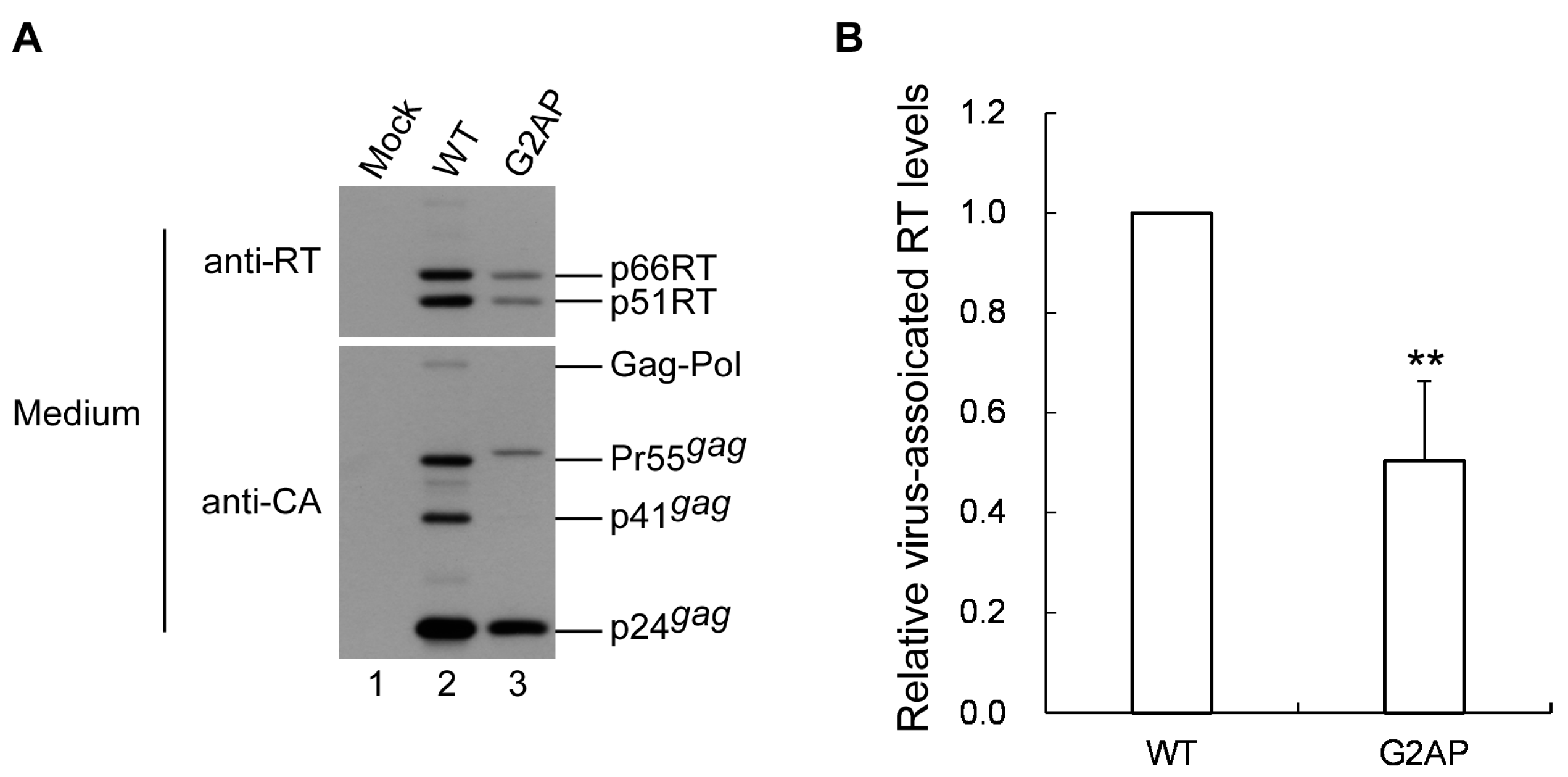
3.6. G2AP Is Defective in Pol Incorporation
3.7. 2A exerts no Major Effects on Post-Assembly Post-Processing Stages of Virus Infectivity
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Petropoulos, C. Retroviral Taxonomy, Protein Structures, Sequences, and Genetic Maps. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997. [Google Scholar]
- Jacks, T.; Power, M.D.; Masiarz, F.R.; Luciw, P.A.; Barr, P.J.; Varmus, H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 1988, 331, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Potempa, M.; Swanstrom, R. The choreography of HIV-1 proteolytic processing and virion assembly. J. Biol. Chem. 2012, 287, 40867–40874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swanstrom, R.; Wills, J.W. Synthesis, Assembly, and Processing of Viral Proteins. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997. [Google Scholar]
- Arrigo, S.J.; Huffman, K. Potent inhibition of human immunodeficiency virus type 1 (HIV-1) replication by inducible expression of HIV-1 PR multimers. J. Virol. 1995, 69, 5988–5994. [Google Scholar] [CrossRef] [Green Version]
- Hill, M.K.; Hooker, C.W.; Harrich, D.; Crowe, S.M.; Mak, J. Gag-Pol supplied in trans is efficiently packaged and supports viral function in human immunodeficiency virus type 1. J. Virol. 2001, 75, 6835–6840. [Google Scholar] [CrossRef] [Green Version]
- Krausslich, H.G. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc. Natl. Acad. Sci. USA 1991, 88, 3213–3217. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Morrow, C.D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 1991, 65, 5111–5117. [Google Scholar] [CrossRef] [Green Version]
- Rose, J.R.; Babe, L.M.; Craik, C.S. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J. Virol. 1995, 69, 2751–2758. [Google Scholar] [CrossRef] [Green Version]
- Shehu-Xhilaga, M.; Crowe, S.M.; Mak, J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001, 75, 1834–1841. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.T.; Chou, Y.C.; Chiang, C.C. Assembly and processing of human immunodeficiency virus Gag mutants containing a partial replacement of the matrix domain by the viral protease domain. J. Virol. 2000, 74, 3418–3422. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Martin, M.A. Incorporation of Pr160(gag-pol) into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J. Virol. 1997, 71, 4472–4478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.J.; Srinivasakumar, N.; Hammarskjöld, M.L.; Rekosh, D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 1993, 67, 2266–2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasakumar, N.; Hammarskjöld, M.L.; Rekosh, D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 1995, 69, 6106–6114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Morrow, C.D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J. Virol. 1992, 66, 6304–6313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.-Z.; Worobey, M. An Endogenous Foamy-like Viral Element in the Coelacanth Genome. PLoS Pathog. 2012, 8, e1002790. [Google Scholar] [CrossRef] [Green Version]
- Löchelt, M.; Flügel, R.M. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J. Virol. 1996, 70, 1033–1040. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.F.; Baldwin, D.N.; Gwynn, S.R.; Yendapalli, S.; Linial, M.L. Human Foamy Virus Replication: A Pathway Distinct from That of Retroviruses and Hepadnaviruses. Science 1996, 271, 1579–1582. [Google Scholar] [CrossRef]
- Cen, S.; Niu, M.; Saadatmand, J.; Guo, F.; Huang, Y.; Nabel, G.J.; Kleiman, L. Incorporation of Pol into Human Immunodeficiency Virus Type 1 Gag Virus-Like Particles Occurs Independently of the Upstream Gag Domain in Gag-Pol. J. Virol. 2004, 78, 1042–1049. [Google Scholar] [CrossRef] [Green Version]
- Chiu, H.-C.; Yao, S.-Y.; Wang, C.-T. Coding Sequences Upstream of the Human Immunodeficiency Virus Type 1 Reverse Transcriptase Domain in Gag-Pol Are Not Essential for Incorporation of the Pr160gag-pol into Virus Particles. J. Virol. 2002, 76, 3221–3231. [Google Scholar] [CrossRef] [Green Version]
- Buchschacher, G.L.; Yu, L.; Murai, F.; Friedmann, T.; Miyanohara, A. Association of Murine Leukemia Virus Pol with Virions, Independent of Gag-Pol Expression. J. Virol. 1999, 73, 9632–9637. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Lee, S.-R.; Li, L.-H.; Park, H.-J.; Park, J.-H.; Lee, K.Y.; Kim, M.-K.; Shin, B.A.; Choi, S.-Y. High Cleavage Efficiency of a 2A Peptide Derived from Porcine Teschovirus-1 in Human Cell Lines, Zebrafish and Mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.-W.; Yu, F.-H.; Huang, K.-J.; Wang, C.-T. P6gag domain confers cis HIV-1 Gag-Pol assembly and release capability. J. Gen. Virol. 2016, 97, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Doyon, L.; Payant, C.; Brakier-Gingras, L.; Lamarre, D. Novel Gag-Pol Frameshift Site in Human Immunodeficiency Virus Type 1 Variants Resistant to Protease Inhibitors. J. Virol. 1998, 72, 6146–6150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.-H.; Huang, K.-J.; Wang, C.-T. C-Terminal HIV-1 Transframe p6* Tetrapeptide Blocks Enhanced Gag Cleavage Incurred by Leucine Zipper Replacement of a Deleted p6* Domain. J. Virol. 2017, 91, e00103–e00117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.-H.; Chou, T.-A.; Liao, W.-H.; Huang, K.-J.; Wang, C.-T. Gag-Pol Transframe Domain p6* Is Essential for HIV-1 Protease-Mediated Virus Maturation. PLoS ONE 2015, 10, e0127974. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.C.; Friedmann, T.; Driever, W.; Burrascano, M.; Yee, J. Vesicular Stomatitis Virus G Glycoprotein Pseudotyped Retroviral Vectors: Concentration to Very High Titer and Efficient Gene Transfer into Mammalian and Nonmammalian Cells. Proc. Natl. Acad. Sci. USA 1993, 90, 8033–8037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferris, A.L.; Hizi, A.; Showalter, S.D.; Pichuantes, S.; Babe, L.; Craik, C.S.; Hughes, S.H. Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure. Virology 1990, 175, 456–464. [Google Scholar] [CrossRef] [Green Version]
- Hizi, A.; McGill, C.; Hughes, S.H. Expression of Soluble, Enzymatically Active, Human Immunodeficiency Virus Reverse Transcriptase in Escherichia coli and Analysis of Mutants. Proc. Natl. Acad. Sci. USA 1988, 85, 1218–1222. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, M.L.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- Donnelly, M.L.L.; Hughes, L.E.; Luke, G.; Mendoza, H.; ten Dam, E.; Gani, D.; Ryan, M.D. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 2001, 82, 1027–1041. [Google Scholar] [CrossRef]
- Cherry, E.; Liang, C.; Rong, L.; Quan, Y.; Inouye, P.; Li, X.; Morin, N.; Kotler, M.; Wainberg, M.A. Characterization of human immunodeficiency virus type-1 (HIV-1) particles that express protease-reverse transcriptase fusion proteins11Edited by J. Karn. J. Mol. Biol. 1998, 284, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Pfrepper, K.-I.; Rackwitz, H.-R.; Schnölzer, M.; Heid, H.; Löchelt, M.; Flügel, R.M. Molecular Characterization of Proteolytic Processing of the Pol Proteins of Human Foamy Virus Reveals Novel Features of the Viral Protease. J. Virol. 1998, 72, 7648–7652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueiredo, A.; Moore, K.L.; Mak, J.; Sluis-Cremer, N.; de Bethune, M.P.; Tachedjian, G. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog. 2006, 2, e119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tachedjian, G.; Orlova, M.; Sarafianos, S.G.; Arnold, E.; Goff, S.P. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 2001, 98, 7188–7193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freed, E.O. HIV-1 Gag Proteins: Diverse Functions in the Virus Life Cycle. Virology 1998, 251, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnelly, M.L.; Gani, D.; Flint, M.; Monaghan, S.; Ryan, M.D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J. Gen. Virol. 1997, 78, 13–21. [Google Scholar] [CrossRef]
- Groot Bramel-Verheije, M.H.; Rottier, P.J.M.; Meulenberg, J.J.M. Expression of a Foreign Epitope by Porcine Reproductive and Respiratory Syndrome Virus. Virology 2000, 278, 380–389. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.H.; Wang, C.T. Characterization of human immunodeficiency virus type 1 Pr160 gag-pol mutants with truncations downstream of the protease domain. Virology 2004, 329, 180–188. [Google Scholar] [CrossRef]
- Abbink, T.E.M.; Ooms, M.; Haasnoot, P.C.J.; Berkhout, B. The HIV-1 Leader RNA Conformational Switch Regulates RNA Dimerization but Does Not Regulate mRNA Translation. Biochemistry 2005, 44, 9058–9066. [Google Scholar] [CrossRef]
- Clever, J.L.; Miranda, J.D.; Parslow, T.G. RNA Structure and Packaging Signals in the 5′ Leader Region of the Human Immunodeficiency Virus Type 1 Genome. J. Virol. 2002, 76, 12381–12387. [Google Scholar] [CrossRef] [Green Version]
- Clever, J.L.; Taplitz, R.A.; Lochrie, M.A.; Polisky, B.; Parslow, T.G. A Heterologous, High-Affinity RNA Ligand for Human Immunodeficiency Virus Gag Protein Has RNA Packaging Activity. J. Virol. 2000, 74, 541–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.; Heng, X.; Summers, M.F. Structural Determinants and Mechanism of HIV-1 Genome Packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lever, A.; Gottlinger, H.; Haseltine, W.; Sodroski, J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luban, J.; Goff, S.P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 1994, 68, 3784–3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBride, M.S.; Panganiban, A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996, 70, 2963–2973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamanian, M.; Purzycka, K.J.; Wille, P.T.; Ha, J.S.; McDonald, D.; Gao, Y.; Le Grice, S.F.J.; Arts, E.J. A cis-Acting Element in Retroviral Genomic RNA Links Gag-Pol Ribosomal Frameshifting to Selective Viral RNA Encapsidation. Cell Host Microbe 2013, 13, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Nikolaitchik, O.A.; Hu, W.-S. Deciphering the Role of the Gag-Pol Ribosomal Frameshift Signal in HIV-1 RNA Genome Packaging. J. Virol. 2014, 88, 4040–4046. [Google Scholar] [CrossRef] [Green Version]
- Comas-Garcia, M.; Datta, S.A.K.; Baker, L.; Varma, R.; Gudla, P.R.; Rein, A. Dissection of specific binding of HIV-1 Gag to the ‘packaging signal’ in viral RNA. elife 2017, 6, e27055. [Google Scholar] [CrossRef]
- Lee, E.-G.; Sinicrope, A.; Jackson, D.L.; Yu, S.F.; Linial, M.L. Foamy Virus Pol Protein Expressed as a Gag-Pol Fusion Retains Enzymatic Activities, Allowing for Infectious Virus Production. J. Virol. 2012, 86, 5992–6001. [Google Scholar] [CrossRef] [Green Version]
- Swiersy, A.; Wiek, C.; Reh, J.; Zentgraf, H.; Lindemann, D. Orthoretroviral-like prototype foamy virus gag-pol expression is compatible with viral replication. Retrovirology 2011, 8, 66. [Google Scholar] [CrossRef] [Green Version]
- Chiu, H.-C.; Wang, F.-D.; Chen, Y.-M.A.; Wang, C.-T. Effects of human immunodeficiency virus type 1 transframe protein p6* mutations on viral protease-mediated Gag processing. J. Gen. Virol. 2006, 87, 2041–2046. [Google Scholar] [CrossRef] [PubMed]





| Construct a | Titer (c.f.u./mL) | Relative Titer | Mean Relative Titer | |
|---|---|---|---|---|
| Mutant | Dp6*PR b | (%) | (%) ± SD c | |
| D2APol | 9 | 5500 | 0.164 | |
| 11 | 5000 | 0.220 | ||
| 8 | 3750 | 0.213 | ||
| 11 | 5500 | 0.200 | 0.199 ± 0.025 | |
| D2A/P22APol | 11,000 | 5500 | 200.0 | |
| 11,000 | 5000 | 220.0 | ||
| 9000 | 3750 | 240.0 | ||
| 8750 | 5500 | 159.1 | 204.8 ± 34.56 | |
| D2A/D15EPol | 8250 | 5500 | 150.0 | |
| 10,500 | 5000 | 210.0 | ||
| 6250 | 3750 | 166.7 | ||
| 8250 | 5500 | 150.0 | 169.2 ± 28.33 | |


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, F.-H.; Huang, K.-J.; Wang, C.-T. HIV-1 Mutant Assembly, Processing and Infectivity Expresses Pol Independent of Gag. Viruses 2020, 12, 54. https://doi.org/10.3390/v12010054
Yu F-H, Huang K-J, Wang C-T. HIV-1 Mutant Assembly, Processing and Infectivity Expresses Pol Independent of Gag. Viruses. 2020; 12(1):54. https://doi.org/10.3390/v12010054
Chicago/Turabian StyleYu, Fu-Hsien, Kuo-Jung Huang, and Chin-Tien Wang. 2020. "HIV-1 Mutant Assembly, Processing and Infectivity Expresses Pol Independent of Gag" Viruses 12, no. 1: 54. https://doi.org/10.3390/v12010054





