The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection
Abstract
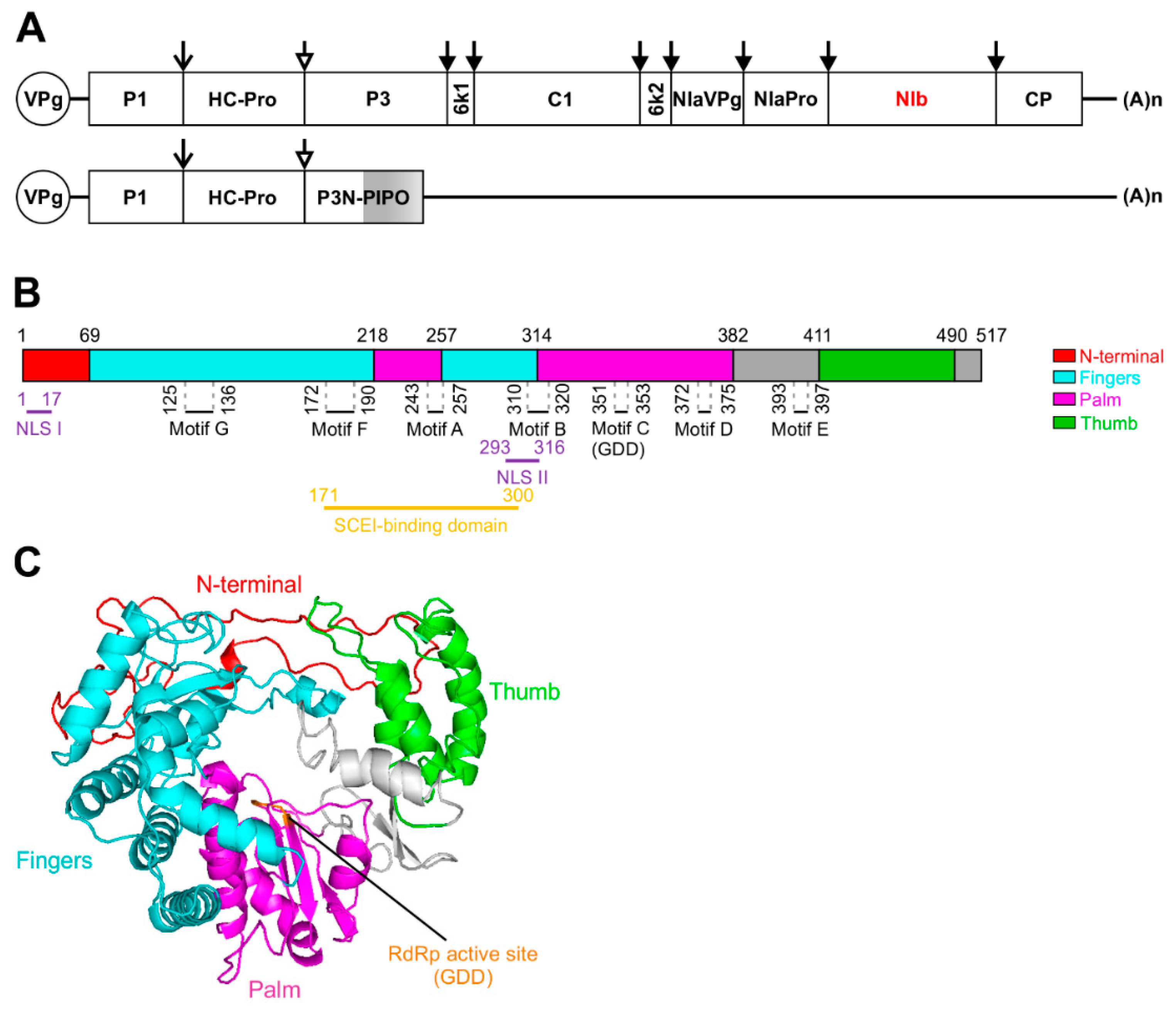
:1. Introduction
2. Structural Characterization of NIb
3. NIb is More Than an RdRp in the VRC
4. NIb and Sumoylation
5. NIb and Autophagy
6. NIb and Pathogenesis
7. Conclusions and Future Prospects
Funding
Acknowledgments
Conflicts of Interest
References
- Cui, H.; Wang, A. The biological impact of the hypervariable N-terminal region of potyvirtal genomes. Annu. Rev. Virol. 2019, 6, 255–274. [Google Scholar] [CrossRef] [PubMed]
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; Lopez-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; Garcia, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar] [PubMed]
- Rybicki, E.P. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Adams, M.J.; Antoniw, J.F.; Beaudoin, F. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 2005, 6, 471–487. [Google Scholar] [CrossRef]
- Chung, B.Y.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [Green Version]
- Hagiwara-Komoda, Y.; Choi, S.H.; Sato, M.; Atsumi, G.; Abe, J.; Fukuda, J.; Honjo, M.N.; Nagano, A.J.; Komoda, K.; Nakahara, K.S.; et al. Truncated yet functional viral protein produced via RNA polymerase slippage implies underestimated coding capacity of RNA viruses. Sci. Rep. 2016, 6, 21411. [Google Scholar] [CrossRef]
- Rodamilans, B.; Valli, A.; Mingot, A.; San Leon, D.; Baulcombe, D.; Lopez-Moya, J.J.; Garcia, J.A. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the potyviridae family. J. Virol. 2015, 89, 6965–6967. [Google Scholar] [CrossRef] [Green Version]
- Mingot, A.; Valli, A.; Rodamilans, B.; San Leon, D.; Baulcombe, D.C.; Garcia, J.A.; Lopez-Moya, J.J. The P1N-PISPO trans-frame gene of sweet potato feathery mottle potyvirus is produced during virus infection and functions as an RNA silencing suppressor. J. Virol. 2016, 90, 3543–3557. [Google Scholar] [CrossRef] [Green Version]
- Olspert, A.; Chung, B.Y.; Atkins, J.F.; Carr, J.P.; Firth, A.E. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. Embo. Rep. 2015, 16, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Valdez, P.; Olvera, R.E.; Carrington, J.C. Functions of the tobacco etch virus RNA polymerase (NIb): Subcellular transport and protein-protein interaction with VPg/proteinase (NIa). J. Virol. 1997, 71, 1598–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mine, A.; Okuno, T. Composition of plant virus RNA replicase complexes. Curr. Opin. Virol. 2012, 2, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Wang, A. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 2008, 82, 12252–12264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Huang, T.S.; McNeil, J.; Laliberte, J.F.; Hong, J.; Nelson, R.S.; Wang, A. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 2010, 84, 799–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M.T.; Gélie, B. Non-structural plum pox potyvirus proteins detected by immunogold labelling. Eur. J. Plant Pathol. 1997, 103, 427–431. [Google Scholar] [CrossRef]
- Restrepo, M.A.; Freed, D.D.; Carrington, J.C. Nuclear transport of plant potyviral proteins. Plant Cell 1990, 2, 987–998. [Google Scholar]
- Ivanov, K.I.; Eskelin, K.; Lohmus, A.; Makinen, K. Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 2014, 95, 1415–1429. [Google Scholar] [CrossRef] [Green Version]
- Grangeon, R.; Cotton, S.; Laliberte, J.F. A model for the biogenesis of turnip mosaic virus replication factories. Commun. Integr. Biol. 2010, 3, 363–365. [Google Scholar] [CrossRef]
- Deng, P.; Wu, Z.; Wang, A. The multifunctional protein CI of potyviruses plays interlinked and distinct roles in viral genome replication and intercellular movement. Virol. J. 2015, 12, 141. [Google Scholar] [CrossRef] [Green Version]
- Hasiow-Jaroszewska, B.; Fares, M.A.; Elena, S.F. Molecular evolution of viral multifunctional proteins: The case of potyvirus HC-Pro. J. Mol. Evol. 2014, 78, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Laliberte, J.F. The genome-linked protein VPg of plant viruses-a protein with many partners. Curr. Opin. Virol. 2011, 1, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Kezar, A.; Kavcic, L.; Polak, M.; Novacek, J.; Gutierrez-Aguirre, I.; Znidaric, M.T.; Coll, A.; Stare, K.; Gruden, K.; Ravnikar, M.; et al. Structural basis for the multitasking nature of the potato virus Y coat protein. Sci. Adv. 2019, 5, eaaw3808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorel, M.; Garcia, J.A.; German-Retana, S. The Potyviridae cylindrical inclusion helicase: A key multipartner and multifunctional protein. Mol. Plant Microbe. Interact. 2014, 27, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Valli, A.A.; Gallo, A.; Rodamilans, B.; Lopez-Moya, J.J.; Garcia, J.A. The HCPro from the Potyviridae family: An enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763. [Google Scholar] [CrossRef] [Green Version]
- Dougherty, W.G.; Cary, S.M.; Parks, T.D. Molecular genetic analysis of a plant virus polyprotein cleavage site: A model. Virology 1989, 171, 356–364. [Google Scholar] [CrossRef]
- Yoon, H.Y.; Hwang, D.C.; Choi, K.Y.; Song, B.D. Proteolytic processing of oligopeptides containing the target sequences by the recombinant tobacco vein mottling virus NIa proteinase. Mol. Cells 2000, 10, 213–219. [Google Scholar] [CrossRef]
- Garcia, J.A.; Lain, S.; Cervera, M.T.; Riechmann, J.L.; Martin, M.T. Mutational analysis of plum pox potyvirus polyprotein processing by the NIa protease in Escherichia coli. J. Gen. Virol. 1990, 71, 2773–2779. [Google Scholar] [CrossRef]
- Garcia, J.A.; Martin, M.T.; Cervera, M.T.; Riechmann, J.L. Proteolytic processing of the plum pox potyvirus polyprotein by the NIa protease at a novel cleavage site. Virology 1992, 188, 697–703. [Google Scholar] [CrossRef]
- Carrington, J.C.; Dougherty, W.G. A viral cleavage site cassette: Identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc. Natl. Acad. Sci. USA 1988, 85, 3391–3395. [Google Scholar] [CrossRef] [Green Version]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origin and evolution of the global RNA virome. mBi 2018, 9, e02329-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Farias, S.T.; Dos Santos Junior, A.P.; Rego, T.G.; Jose, M.V. Origin and evolution of RNA-dependent RNA polymerase. Front. Genet. 2017, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, E.K.; Kao, C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 1998, 252, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Bruenn, J.A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003, 31, 1821–1829. [Google Scholar] [CrossRef]
- Ferrer-Orta, C.; Arias, A.; Escarmis, C.; Verdaguer, N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006, 16, 27–34. [Google Scholar] [CrossRef]
- Shatskaya, G.S.; Drutsa, V.L.; Koroleva, O.N.; Osterman, I.A.; Dmitrieva, T.M. Investigation of activity of recombinant mengovirus RNA-dependent RNA polymerase and its mutants. Biochemistry 2013, 78, 96–101. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479–480, 2–25. [Google Scholar] [CrossRef] [Green Version]
- Ferrer-Orta, C.; Ferrero, D.; Verdaguer, N. RNA-dependent RNA polymerases of picornaviruses: From the structure to regulatory mechanisms. Viruses 2015, 7, 4438–4460. [Google Scholar] [CrossRef] [Green Version]
- Garriga, D.; Ferrer-Orta, C.; Querol-Audi, J.; Oliva, B.; Verdaguer, N. Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 2013, 425, 2279–2287. [Google Scholar] [CrossRef] [Green Version]
- Vives-Adrian, L.; Lujan, C.; Oliva, B.; van der Linden, L.; Selisko, B.; Coutard, B.; Canard, B.; van Kuppeveld, F.J.; Ferrer-Orta, C.; Verdaguer, N. The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site. J. Virol. 2014, 88, 5595–5607. [Google Scholar] [CrossRef] [Green Version]
- Moradi, Z.; Mehrvar, M.; Nazifi, E.; Zakiaghl, M. Iranian johnsongrass mosaic virus: The complete genome sequence, molecular and biological characterization, and comparison of coat protein gene sequences. Virus Genes 2017, 53, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Worrall, E.A.; Hayward, A.C.; Fletcher, S.J.; Mitter, N. Molecular characterization and analysis of conserved potyviral motifs in bean common mosaic virus (BCMV) for RNAi-mediated protection. Arch. Virol. 2019, 164, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wayper, P.J.; Gibbs, A.J.; Fourment, M.; Rodoni, B.C.; Gibbs, M.J. Accumulating variation at conserved sites in potyvirus genomes is driven by species discovery and affects degenerate primer design. PLoS ONE 2008, 3, e1586. [Google Scholar] [CrossRef] [PubMed]
- Allison, R.; Johnston, R.E.; Dougherty, W.G. The nucleotide sequence of the coding region of tobacco etch virus genomic RNA: Evidence for the synthesis of a single polyprotein. Virology 1986, 154, 9–20. [Google Scholar] [CrossRef]
- Li, X.H.; Carrington, J.C. Complementation of tobacco etch potyvirus mutants by active RNA polymerase expressed in transgenic cells. Proc. Natl. Acad. Sci. USA 1995, 92, 457–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majer, E.; Salvador, Z.; Zwart, M.P.; Willemsen, A.; Elena, S.F.; Daros, J.A. Relocation of the NIb gene in the tobacco etch potyvirus genome. J. Virol. 2014, 88, 4586–4590. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Shi, Y.; Luo, Z.; Lu, Y.; Zheng, H.; Yan, F.; Chen, J.; Chen, J.; Adams, M.J.; Wu, Y. Protein-protein interactions in two potyviruses using the yeast two-hybrid system. Virus Res. 2009, 142, 36–40. [Google Scholar] [CrossRef]
- Yambao, M.L.; Masuta, C.; Nakahara, K.; Uyeda, I. The central and C-terminal domains of VPg of Clover yellow vein virus are important for VPg-HCPro and VPg-VPg interactions. J. Gen. Virol. 2003, 84, 2861–2869. [Google Scholar] [CrossRef]
- Chinnaswamy, S.; Murali, A.; Li, P.; Fujisaki, K.; Kao, C.C. Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J. Virol. 2010, 84, 5923–5935. [Google Scholar] [CrossRef] [Green Version]
- Hogbom, M.; Jager, K.; Robel, I.; Unge, T.; Rohayem, J. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J. Gen. Virol. 2009, 90, 281–291. [Google Scholar] [CrossRef]
- Kaiser, W.J.; Chaudhry, Y.; Sosnovtsev, S.V.; Goodfellow, I.G. Analysis of protein-protein interactions in the feline calicivirus replication complex. J. Gen. Virol. 2006, 87, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Hamatake, R.K.; Mathis, D.M.; Racela, J.; Rigat, K.L.; Lemm, J.; Colonno, R.J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 2000, 74, 851–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cevik, B. The RNA-dependent RNA polymerase of Citrus tristeza virus forms oligomers. Virology 2013, 447, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberte, J.F. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanfaçon, H. Replication of positive-strand RNA viruses in plants: Contact points between plant and virus components. Can. J. Bot. 2005, 83, 1529–1549. [Google Scholar] [CrossRef]
- Gonzalez, R.; Wu, B.; Li, X.; Martinez, F.; Elena, S.F. Mutagenesis scanning uncovers evolutionary constraints on tobacco etch potyvirus membrane-associated 6K2 protein. Genome Biol. Evol. 2019, 11, 1207–1222. [Google Scholar] [CrossRef] [Green Version]
- Patarroyo, C.; Laliberte, J.F.; Zheng, H. Hijack it, change it: How do plant viruses utilize the host secretory pathway for efficient viral replication and spread? Front. Plant Sci. 2012, 3, 308. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Patarroyo, C.; Garcia Cabanillas, D.; Zheng, H.; Laliberte, J.F. The vesicle-forming 6K2 protein of turnip mosaic virus interacts with the COPII coatomer Sec24a for viral systemic infection. J. Virol. 2015, 89, 6695–6710. [Google Scholar] [CrossRef] [Green Version]
- Movahed, N.; Sun, J.; Vali, H.; Laliberte, J.F.; Zheng, H. A host ER fusogen is recruited by turnip mosaic virus for maturation of viral replication vesicles. Plant Physiol. 2019, 179, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Zhang, C.; Hou, X.; Sanfacon, H.; Wang, A. The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog. 2013, 9, e1003378. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Basu, K.; Mui, J.; Vali, H.; Zheng, H.; Laliberte, J.F. Ultrastructural characterization of turnip mosaic virus-induced cellular rearrangements reveals membrane-bound viral particles accumulating in vacuoles. J. Virol. 2015, 89, 12441–12456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, X.; Cao, X.; Wang, X.; Jiang, J.; Wan, J.; Laliberte, J.F.; Zhang, Y. Three-dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front. Plant Sci. 2018, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Yaghmaiean, H.; Wu, G.; Wu, X.; Chen, X.; Thorn, G.; Wang, A. The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex. Virology 2017, 510, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Lohmus, A.; Varjosalo, M.; Makinen, K. Protein composition of 6K2-induced membrane structures formed during Potato virus A infection. Mol. Plant Pathol. 2016, 17, 943–958. [Google Scholar] [CrossRef] [Green Version]
- Daros, J.A.; Schaad, M.C.; Carrington, J.C. Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 1999, 73, 8732–8740. [Google Scholar] [CrossRef] [Green Version]
- Fellers, J.; Wan, J.; Hong, Y.; Collins, G.B.; Hunt, A.G. In vitro interactions between a potyvirus-encoded, genome-linked protein and RNA-dependent RNA polymerase. J. Gen. Virol. 1998, 79 Pt 8, 2043–2949. [Google Scholar] [CrossRef]
- Haldeman-Cahill, R.; Daros, J.A.; Carrington, J.C. Secondary structures in the capsid protein coding sequence and 3’ nontranslated region involved in amplification of the tobacco etch virus genome. J. Virol. 1998, 72, 4072–4079. [Google Scholar] [CrossRef] [Green Version]
- Makinen, K.; Hafren, A. Intracellular coordination of potyviral RNA functions in infection. Front. Plant Sci. 2014, 5, 110. [Google Scholar] [CrossRef] [Green Version]
- Anindya, R.; Chittori, S.; Savithri, H.S. Tyrosine 66 of Pepper vein banding virus genome-linked protein is uridylylated by RNA-dependent RNA polymerase. Virology 2005, 336, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Puustinen, P.; Makinen, K. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 2004, 279, 38103–38110. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ullah, Z.; Grumet, R. Interaction between zucchini yellow mosaic potyvirus RNA-dependent RNA polymerase and host poly-(A) binding protein. Virology 2000, 275, 433–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufresne, P.J.; Thivierge, K.; Cotton, S.; Beauchemin, C.; Ide, C.; Ubalijoro, E.; Laliberte, J.F.; Fortin, M.G. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 2008, 374, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufresne, P.J.; Ubalijoro, E.; Fortin, M.G.; Laliberte, J.F. Arabidopsis thaliana class II poly(A)-binding proteins are required for efficient multiplication of turnip mosaic virus. J. Gen. Virol. 2008, 89, 2339–2348. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Shine, M.B.; Cui, X.; Chen, X.; Ma, N.; Kachroo, P.; Zhi, H.; Kachroo, A. The Potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef] [Green Version]
- Thivierge, K.; Cotton, S.; Dufresne, P.J.; Mathieu, I.; Beauchemin, C.; Ide, C.; Fortin, M.G.; Laliberte, J.F. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 2008, 377, 216–225. [Google Scholar] [CrossRef]
- Huang, T.S.; Wei, T.; Laliberté, J.-F.; Wang, A. A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol. 2010, 152, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Pogany, J.; Tupman, S.; Esposito, A.M.; Kinzy, T.G.; Nagy, P.D. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 2010, 6, e1001175. [Google Scholar] [CrossRef] [Green Version]
- Mine, A.; Hyodo, K.; Tajima, Y.; Kusumanegara, K.; Taniguchi, T.; Kaido, M.; Mise, K.; Taniguchi, H.; Okuno, T. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J. Virol. 2012, 86, 12091–12104. [Google Scholar] [CrossRef] [Green Version]
- Beauchemin, C.; Boutet, N.; Laliberte, J.F. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J. Virol. 2007, 81, 775–782. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xiong, R.; Bernards, M.; Wang, A. Recruitment of arabidopsis RNA helicase AtRH9 to the viral replication complex by viral replicase to promote turnip mosaic virus replication. Sci. Rep. 2016, 6, 30297. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Li, F.; Renaud, J.; Shen, W.; Li, Y.; Guo, L.; Cui, H.; Sumarah, M.; Wang, A. NbEXPA1, an alpha-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. Plant J. 2017, 92, 846–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcaide-Loridan, C.; Jupin, I. Ubiquitin and plant viruses, let’s play together! Plant Physiol. 2012, 160, 72–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 2015, 53, 45–66. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, A. Multifaceted defense and counter-defense in co-evolutionary arms race between plants and viruses. Commun. Integr. Biol. 2017, 10, e1341025. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Xiong, R.; Li, Y.; Li, F.; Zhou, X.; Wang, A. Sumoylation of turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 2017, 29, 508–525. [Google Scholar] [CrossRef] [Green Version]
- Xiong, R.; Wang, A. SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection. J. Virol. 2013, 87, 4704–4715. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.; Withers, J.; Mohan, R.; Marques, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe. 2015, 18, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Detchemendy, T.W.; Pajerowska-Mukhtar, K.M.; Mukhtar, M.S. NPR1 in JazzSet with pathogen effectors. Trends Plant Sci. 2018, 23, 469–472. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Despres, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Mohan, R.; Zhang, Y.; Li, M.; Chen, H.; Palmer, I.A.; Chang, M.; Qi, G.; Spoel, S.H.; Mengiste, T.; et al. NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 2019, 181, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Bassham, D.C. Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol. 2018, 8, 180162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Mugume, Y.; Bassham, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife 2017, 6, e23897. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.S.; Masuta, C.; Yamada, S.; Shimura, H.; Kashihara, Y.; Wada, T.S.; Meguro, A.; Goto, K.; Tadamura, K.; Sueda, K.; et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 2012, 109, 10113–10118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Zhang, C.; Li, Y.; Wu, G.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1268. [Google Scholar] [CrossRef]
- Hafren, A.; Ustun, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 2018, 176, 649–662. [Google Scholar] [CrossRef] [Green Version]
- Kushwaha, N.K.; Hafren, A.; Hofius, D. Autophagy-virus interplay in plants: From antiviral recognition to proviral manipulation. Mol. Plant Pathol. 2019, 20, 1211–1216. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Wang, A. The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 2017, 91, e01478-16. [Google Scholar] [CrossRef] [Green Version]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudel, D.B.; Sanfacon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Valli, A.; Garcia, J.A.; Zhou, X.; Cheng, X. The tug-of-war between plants and viruses: Great progress and many remaining questions. Viruses 2019, 11, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- De Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef] [Green Version]
- Gouveia, B.C.; Calil, I.P.; Machado, J.P.; Santos, A.A.; Fontes, E.P. Immune receptors and co-receptors in antiviral innate immunity in plants. Front. Microbiol. 2016, 7, 2139. [Google Scholar] [CrossRef] [Green Version]
- Maule, A.J.; Caranta, C.; Boulton, M.I. Sources of natural resistance to plant viruses: Status and prospects. Mol. Plant Pathol. 2007, 8, 223–231. [Google Scholar] [CrossRef]
- Janzac, B.F.; Fabre, M.F.; Palloix, A.; Moury, B. Phenotype and spectrum of action of the Pvr4 resistance in pepper against potyviruses, and selection for virulent variants. Plant Pathol. 2009, 58, 443–449. [Google Scholar] [CrossRef]
- Janzac, B.; Montarry, J.; Palloix, A.; Navaud, O.; Moury, B. A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol. Plant Microbe. Interact. 2010, 23, 823–830. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.B.; Lee, H.Y.; Seo, S.; Lee, J.H.; Choi, D. RNA-dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS ONE 2015, 10, e0119639. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.B.; Lee, H.Y.; Choi, E.H.; Park, E.; Kim, J.H.; Moon, K.B.; Kim, H.S.; Choi, D. The coiled-coil and leucine-rich repeat domain of the potyvirus resistance protein Pvr4 has a distinct role in signaling and pathogen recognition. Mol. Plant Microbe Interact. 2018, 31, 906–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fellers, J.P.; Tremblay, D.; Handest, M.F.; Lommel, S.A. The Potato virusd Y MSNR NIb-replicase is the elicitor of a veinal necrosis-hypersensitive response in root knot nematode resistant tobacco. Mol. Plant Pathol. 2002, 3, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Janzac, B.; Fabre, F.; Palloix, A.; Moury, B. Constraints on evolution of virus avirulence factors predict the durability of corresponding plant resistances. Mol. Plant Pathol. 2009, 10, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Moury, B.; Desbiez, C.; Jacquemond, M.; Lecoq, H. Genetic diversity of plant virus populations: Towards hypothesis testing in molecular epidemiology. Adv. Virus Res. 2006, 67, 49–87. [Google Scholar]
- Wallis, C.M.; Stone, A.L.; Sherman, D.J.; Damsteegt, V.D.; Gildow, F.E.; Schneider, W.L. Adaption of plum pox virus to be a herbaceous host (Pisum sativum) following serial passages. J. Gen. Virol. 2007, 88, 2839–2845. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, P.; Zhong, K.; Zhang, F.; Xu, M.; He, L.; Jin, P.; Chen, J.; Yang, J. Wheat yello mosaic virus NIb interacting with host light induced protein (LIP) facilitates its infection through perturbing the abscisic acid pathway in wheat. Biology 2019, 8, 80. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Cao, X.; Li, D. Architecture of viral replication factories. Oncotarget 2015, 6, 30439–30440. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, B.; Ding, Z.; Li, G.; Liu, M.; Zhu, D.; Sun, Y.; Dong, S.; Lou, Z. Distinct mechanism for the formation of the ribonucleoprotein complex of tomato spotted wilt virus. J. Virol. 2017, 91, e00892-17. [Google Scholar] [CrossRef] [Green Version]
- Komoda, K.; Narita, M.; Yamashita, K.; Tanaka, I.; Yao, M. Asymmetric trimeric ring structure of the nucleocapsid protein of Tospovirus. J. Virol. 2017, 91, e01002–e01017. [Google Scholar] [CrossRef] [Green Version]
- Huynh, N.T.; Hesketh, E.L.; Saxena, P.; Meshcheriakova, Y.; Ku, Y.C.; Hoang, L.T.; Johnson, J.E.; Ranson, N.A.; Lomonossoff, G.P.; Reddy, V.S. Crystal structure and proteomics analysis of empty virus-like particles of Cowpea mosaic virus. Structure 2016, 24, 567–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, W.; Shi, Y.; Dai, Z.; Wang, A. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses 2020, 12, 77. https://doi.org/10.3390/v12010077
Shen W, Shi Y, Dai Z, Wang A. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses. 2020; 12(1):77. https://doi.org/10.3390/v12010077
Chicago/Turabian StyleShen, Wentao, Yan Shi, Zhaoji Dai, and Aiming Wang. 2020. "The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection" Viruses 12, no. 1: 77. https://doi.org/10.3390/v12010077
APA StyleShen, W., Shi, Y., Dai, Z., & Wang, A. (2020). The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. Viruses, 12(1), 77. https://doi.org/10.3390/v12010077




