Genome Sequence and Characterization of Five Bacteriophages Infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Phage Isolation and Propagation
2.3. Electron Microscopy Observation of Phage Virions
2.4. Phage Infection Curves
2.5. Host Range Determination
2.6. DNA Isolation
2.7. DNA Sequencing and Genome Assembly
2.8. Gene Prediction and Functional Annotation
2.9. Genome Comparison and Classification
2.10. Protein Domain-Based Classification
3. Results
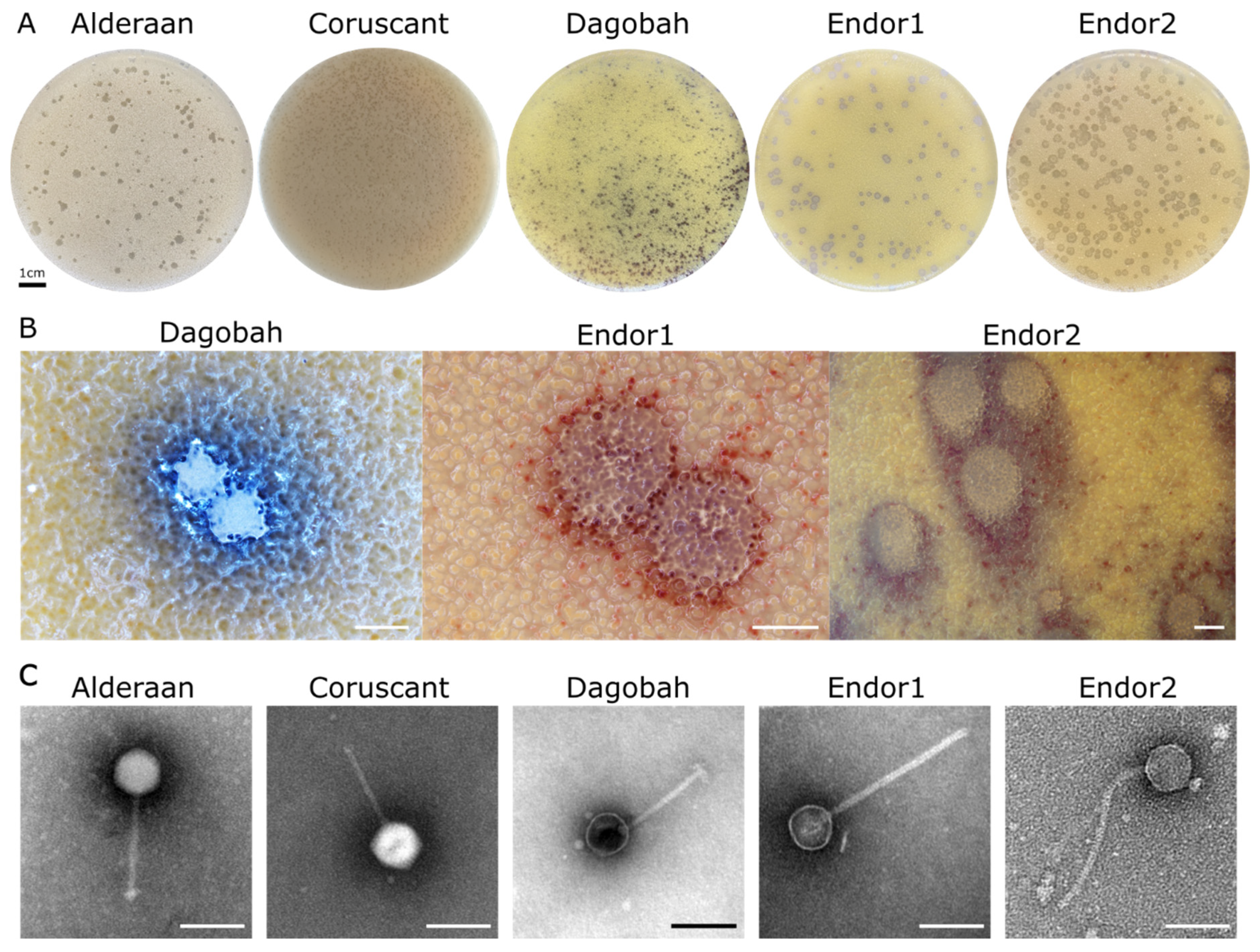
3.1. Phage Isolation and Virion Morphology
3.2. Infection Curves and Host-Range Determination
3.3. Genome Sequencing and Genome Features
3.4. Average Nucleotide Identity (ANI) Analysis
3.5. Protein Domain-Based Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bibb, M.J. Understanding and manipulating antibiotic production in actinomycetes. Biochem. Soc. Trans. 2013, 41, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Streptomyces in Nature and Medicine: The Antibiotic Makers; Oxford University Press: Oxford, NY, USA, 2007; ISBN 978-0-19-515066-7. [Google Scholar]
- Keiser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000; ISBN 0-7084-0623-8. [Google Scholar]
- Elliot, M.A.; Buttner, M.J.; Nodwell, J.R. 24 Multicellular Development in Streptomyces. Myxobacteria 2008, 419–438. [Google Scholar] [CrossRef]
- McCormick, J.R.; Flärdh, K. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 2012, 36, 206–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anne, J.; Wohlleben, W.; Burkardt, H.J.; Springer, R.; Pohler, A. Morphological and Molecular Characterization of Several Actinophages Isolated from Soil Which Lyse Streptomyces cattleya or S. venezuelae. Microbiology 1984, 130, 2639–2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donadio, S.; Paladino, R.; Costanzi, I.; Sparapani, P.; Schreil, W.; Iaccarino, M. Characterization of bacteriophages infecting Streptomyces erythreus and properties of phage-resistant mutants. J. Bacteriol. 1986, 166, 1055–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowding, J.E. Characterization of a Bacteriophage Virulent for Streptomyces coelicolor A3 (2) | Microbiology Society. J. Gen. Microbiol. 1973, 76, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.C.M.; Hendrix, R.W.; Dedrick, R.; Mitchell, K.; Ko, C.-C.; Russell, D.; Bell, E.; Gregory, M.; Bibb, M.J.; Pethick, F.; et al. Evolutionary Relationships among Actinophages and a Putative Adaptation for Growth in Streptomycesspp. J. Bacteriol. 2013, 195, 4924–4935. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.C.M.; Burns, R.N.; Wilson, S.E.; Gregory, M.A. The complete genome sequence of the Streptomyces temperate phage φC31: Evolutionary relationships to other viruses. Nucleic Acids Res. 1999, 27, 2145–2155. [Google Scholar] [CrossRef]
- Lomovskaya, N.D.; Mkrtumian, N.M.; Gostimskaya, N.L.; Danilenko, V.N. Characterization of Temperate Actinophage φC31 Isolated from Streptomyces coelicolor A3(2). J. Virol. 1972, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Lomovskaya, N.D.; Chater, K.F.; Mkrtumian, N.M. Genetics and molecular biology of Streptomyces bacteriophages. Microbiol. Mol. Biol. Rev. 1980, 44, 206–229. [Google Scholar] [CrossRef] [Green Version]
- Burke, J.; Schneider, D.; Westpheling, J. Generalized transduction in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 2001, 98, 6289–6294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosner, A.; Gustein, R. Adsorption of actinophage Pal 6 to developing mycelium of Streptomyces. Can. J. Microbiol. 1981, 27, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.C.; Burnett, S.H.; Carson, S.; Caruso, S.M.; Clase, K.; DeJong, R.J.; Dennehy, J.J.; Denver, D.R.; Dunbar, D.; Elgin, S.C.R.; et al. A Broadly Implementable Research Course in Phage Discovery and Genomics for First-Year Undergraduate Students. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pullan, S.T.; Chandra, G.; Bibb, M.J.; Merrick, M. Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genom. 2011, 12, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, D.; Karoonuthaisiri, N.; Tsai, H.-H.; Huang, C.-H.; Ho, M.-L.; Gai, S.; Patel, K.G.; Huang, J.; Cohen, S.N.; Hopwood, D.A.; et al. Genome plasticity in Streptomyces: Identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome: Identification of 1 Mb TIRs in S. coelicolor A3(2). Mol. Microbiol. 2004, 51, 1535–1550. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Shepherd, M.D.; Kharel, M.K.; Bosserman, M.A.; Rohr, J. Laboratory Maintenance of Streptomyces species. Curr. Protoc. Microbiol. 2010, 18, 10E.1.1–10E.1.8. [Google Scholar] [CrossRef] [Green Version]
- Kauffman, K.M.; Polz, M.F. Streamlining standard bacteriophage methods for higher throughput. MethodsX 2018, 5, 159–172. [Google Scholar] [CrossRef]
- Kensy, F.; Zang, E.; Faulhammer, C.; Tan, R.-K.; Büchs, J. Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb. Cell Fact. 2009, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Rückert, C.; Albersmeier, A.; Busche, T.; Jaenicke, S.; Winkler, A.; Friðjónsson, Ó.H.; Hreggviðsson, G.Ó.; Lambert, C.; Badcock, D.; Bernaerts, K.; et al. Complete genome sequence of Streptomyces lividans TK24. J. Biotechnol. 2015, 199, 21–22. [Google Scholar] [CrossRef]
- Gordon, D.; Green, P. Consed: A graphical editor for next-generation sequencing. Bioinformatics 2013, 29, 2936–2937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Grazziotin, A.L.; Koonin, E.V.; Kristensen, D.M. Prokaryotic Virus Orthologous Groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2017, 45, D491–D498. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [Green Version]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef]
- Tynecki, P.; Guziński, A.; Kazimierczak, J.; Jadczuk, M.; Dastych, J.; Onisko, A. PhageAI—Bacteriophage Life Cycle Recognition with Machine Learning and Natural Language Processing. Bioinformatics 2020. [Google Scholar] [CrossRef]
- Russell, D.A.; Hatfull, G.F. PhagesDB: The actinobacteriophage database. Bioinformatics 2017, 33, 784–786. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, L. Widdowquinn/pyani. 2020. Available online: https://github.com/widdowquinn (accessed on 21 September 2020).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Rudd, B.A.M.; Hopwood, D.A. Genetics of Actinorhodin Biosynthesis by Streptomyces coelicolor A3(2). Microbiology 1979, 114, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Abedon, S.T. Detection of Bacteriophages: Phage Plaques. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–32. ISBN 978-3-319-40598-8. [Google Scholar]
- Botstein, D. A Theory of Modular Evolution for Bacteriophages. Ann. N. Y. Acad. Sci. 1980, 354, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H.; Desiere, F. Comparative phage genomics and the evolution of Siphoviridae: Insights from dairy phages. Mol. Microbiol. 2001, 39, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Molle, V.; Palframan, W.J.; Findlay, K.C.; Buttner, M.J. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 2000, 182, 1286–1295. [Google Scholar] [CrossRef] [Green Version]
- van Wezel, G.P.; van der Meulen, J.; Kawamoto, S.; Luiten, R.G.M.; Koerten, H.K.; Kraal, B. ssgA Is Essential for Sporulation of Streptomyces coelicolor A3(2) and Affects Hyphal Development by Stimulating Septum Formation. J. Bacteriol. 2000, 182, 5653–5662. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, E.; Hünnefeld, M.; Popa, O.; Frunzke, J. Impact of Xenogeneic Silencing on Phage–Host Interactions. J. Mol. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S.T. Chapter 7—Bacteriophage Host Range and Bacterial Resistance. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2010; Volume 70, pp. 217–248. [Google Scholar]
- Morris, P.; Marinelli, L.J.; Jacobs-Sera, D.; Hendrix, R.W.; Hatfull, G.F. Genomic Characterization of Mycobacteriophage Giles: Evidence for Phage Acquisition of Host DNA by Illegitimate Recombination. J. Bacteriol. 2008, 190, 2172–2182. [Google Scholar] [CrossRef] [Green Version]
- Rybniker, J.; Nowag, A.; van Gumpel, E.; Nissen, N.; Robinson, N.; Plum, G.; Hartmann, P. Insights into the function of the WhiB-like protein of mycobacteriophage TM4—A transcriptional inhibitor of WhiB2. Mol. Microbiol. 2010, 77, 642–657. [Google Scholar] [CrossRef]
- Van Dessel, W.; Van Mellaert, L.; Liesegang, H.; Raasch, C.; DeKeersmaeker, S.; Geukens, N.; Lammertyn, E.; Streit, W.; Anné, J. Complete genomic nucleotide sequence and analysis of the temperate bacteriophage VWB. Virology 2005, 331, 325–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehrke, E.J.; Zhang, X.; Pimentel-Elardo, S.M.; Johnson, A.R.; Rees, C.A.; Jones, S.E.; Gehrke, S.S.; Turvey, S.; Boursalie, S.; Hill, J.E.; et al. Silencing cryptic specialized metabolism in Streptomyces by the nucleoid-associated protein Lsr2. eLife 2019, 8, e47691. [Google Scholar] [CrossRef]
- Pfeifer, E.; Hünnefeld, M.; Popa, O.; Polen, T.; Kohlheyer, D.; Baumgart, M.; Frunzke, J. Silencing of cryptic prophages in Corynebacterium glutamicum. Nucleic Acids Res. 2016, 44, 10117–10131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmour, C.M.; Noller, E.C.; Watkins, B. Studies on Streptomyces Phage: I. Growth Characteristics of the Streptomyces griseus Host-Phage System. J. Bacteriol. 1959, 78, 186–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.D.; Kalachová, L.; Novotná, J.; Holub, M.; Kofroňová, O.; Benada, O.; Thompson, C.J.; Weiser, J. Cultivation System Using Glass Beads Immersed in Liquid Medium Facilitates Studies of Streptomyces Differentiation. Appl. Environ. Microbiol. 2005, 71, 2848–2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chater, K. David Hopwood and the emergence of Streptomyces genetics. Int. Microbiol. 1999, 2, 61–68. [Google Scholar]
- Kronheim, S.; Daniel-Ivad, M.; Duan, Z.; Hwang, S.; Wong, A.I.; Mantel, I.; Nodwell, J.R.; Maxwell, K.L. A chemical defence against phage infection. Nature 2018, 564, 283. [Google Scholar] [CrossRef]




| Alderaan | Coruscant | Dagobah | Endor1 | Endor2 | |
|---|---|---|---|---|---|
| S. venezuelae | |||||
| S. coelicolor M600 | |||||
| S. coelicolor M145 | 1 | 1 | 1 | ||
| S. rimosus subsp. rimosus | |||||
| S. scabiei | |||||
| S. griseus | |||||
| S. platensis | |||||
| S. xanthochromogenes | |||||
| S. lividans | 0.2 | ||||
| S. olivaceus | 4 | ||||
| S. cyaneofuscatus | 0.08 | 0.4 |
| Phage Name | Accession Number | Reference Host | Genome Size (kb) | GC Content (%) | ORF Number | Genome Termini Class | Lifestyle Prediction |
|---|---|---|---|---|---|---|---|
| Alderaan | MT711975 | Streptomyces venezuelae ATCC 10712 | 39 | 72.1 | 51 | Headful (pac) | Temperate |
| Coruscant | MT711976 | Streptomyces venezuelae ATCC 10712 | 133 (12 kb DTR) | 48.4 | 290 | DTR (long) | Virulent |
| Dagobah | MT711977 | Streptomyces coelicolor M600 | 47 kb (1 kb DTR) | 68.9 | 93 | DTR (short) | Temperate |
| Endor1 | MT711978 | Streptomyces coelicolor M600 | 49 | 65.8 | 75 | Headful (pac) | Temperate |
| Endor2 | MT711979 | Streptomyces coelicolor M600 | 48 | 65.1 | 75 | Headful (pac) | Temperate |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hardy, A.; Sharma, V.; Kever, L.; Frunzke, J. Genome Sequence and Characterization of Five Bacteriophages Infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2. Viruses 2020, 12, 1065. https://doi.org/10.3390/v12101065
Hardy A, Sharma V, Kever L, Frunzke J. Genome Sequence and Characterization of Five Bacteriophages Infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2. Viruses. 2020; 12(10):1065. https://doi.org/10.3390/v12101065
Chicago/Turabian StyleHardy, Aël, Vikas Sharma, Larissa Kever, and Julia Frunzke. 2020. "Genome Sequence and Characterization of Five Bacteriophages Infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2" Viruses 12, no. 10: 1065. https://doi.org/10.3390/v12101065
APA StyleHardy, A., Sharma, V., Kever, L., & Frunzke, J. (2020). Genome Sequence and Characterization of Five Bacteriophages Infecting Streptomyces coelicolor and Streptomyces venezuelae: Alderaan, Coruscant, Dagobah, Endor1 and Endor2. Viruses, 12(10), 1065. https://doi.org/10.3390/v12101065





