p24G1 Encoded by Grapevine Leafroll-Associated Virus 1 Suppresses RNA Silencing and Elicits Hypersensitive Response-Like Necrosis in Nicotiana Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions, and Preparation of Plasmids
2.2. Agroinfiltration and Fluorescence Imaging
2.3. Northern Blot
2.4. Protein Interaction Analysis in Yeast
2.5. Protein Expression and Purification
2.6. Plant Protein Extraction and Western Blot Analysis
2.7. Electrophoretic Mobility Shift Assay (EMSA)
2.8. Cell Death Analysis and H2O2 Detection
2.9. RT-PCR, qRT-PCR and Statistical Analysis
3. Results
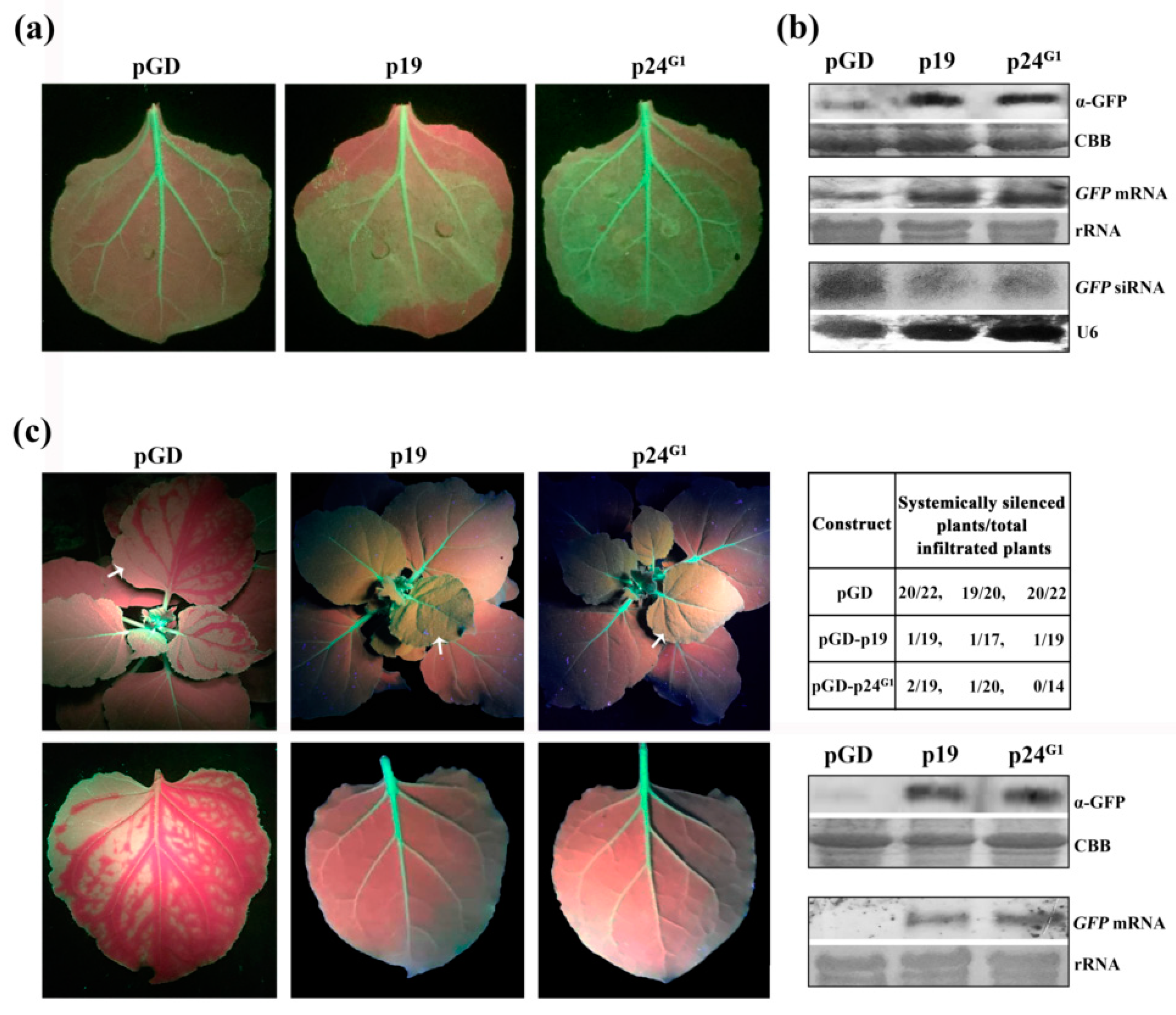
3.1. p24G1 Suppresses Local and Systemic RNA Silencing
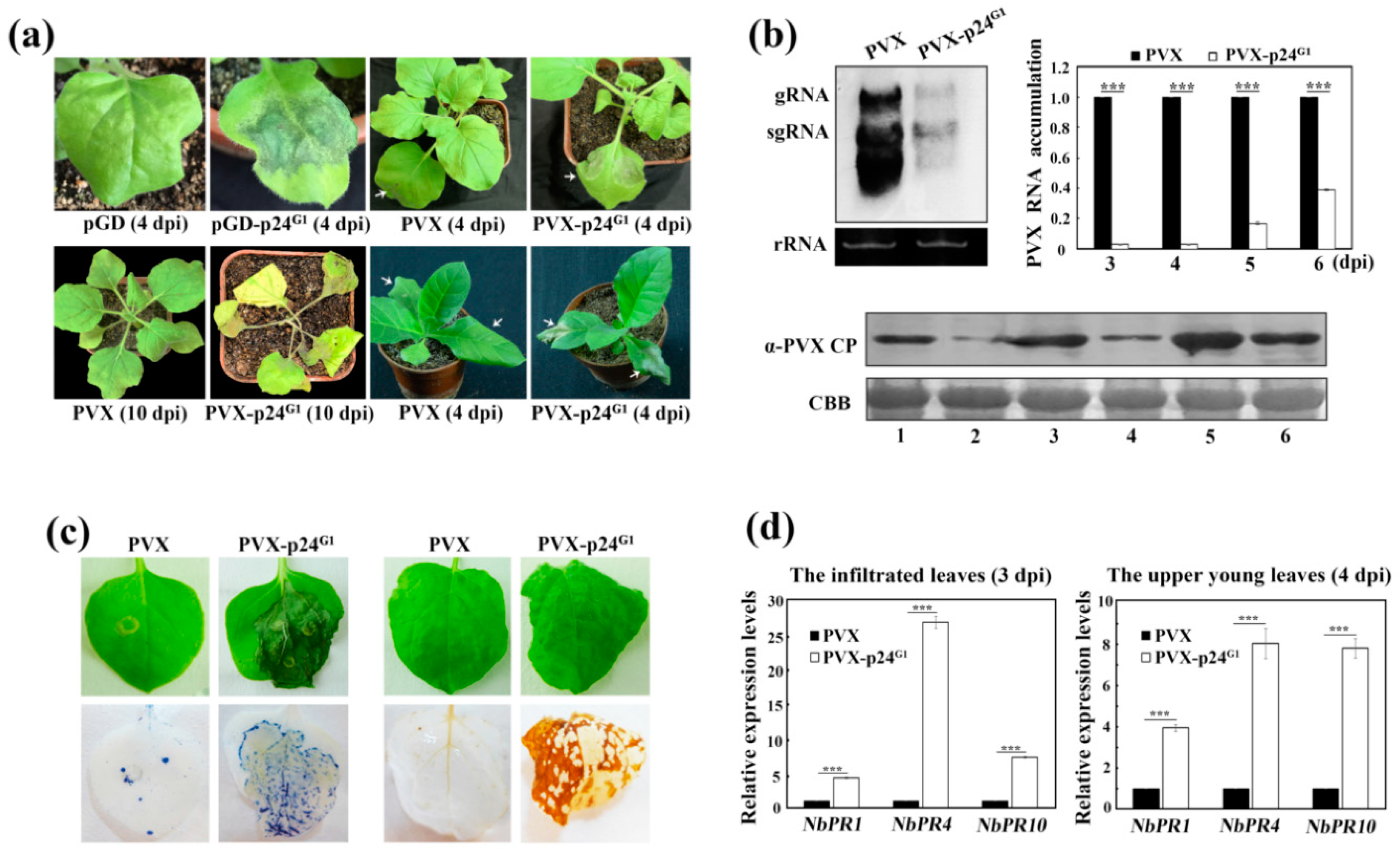
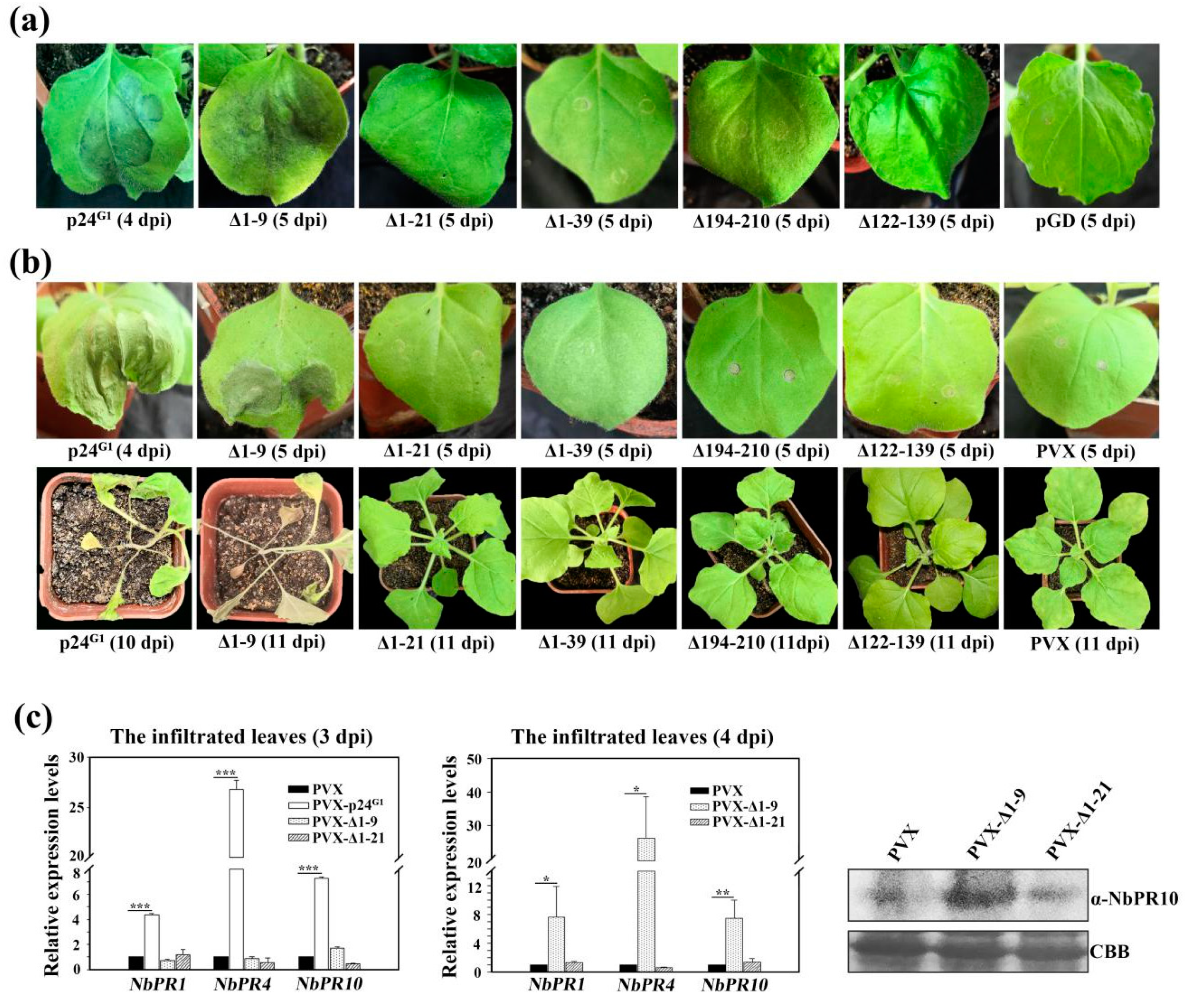
3.2. p24G1 Is a Factor in Pathogenesis Eliciting HR-Like Necrosis in Nicotiana Species
3.3. p24G1 Self-Interacts in the Nucleus Through Its 10–210 aa Region
3.4. Dimerization of p24G1 Is a Prerequisite for Its Nuclear Targeting Mediated by Importin α1
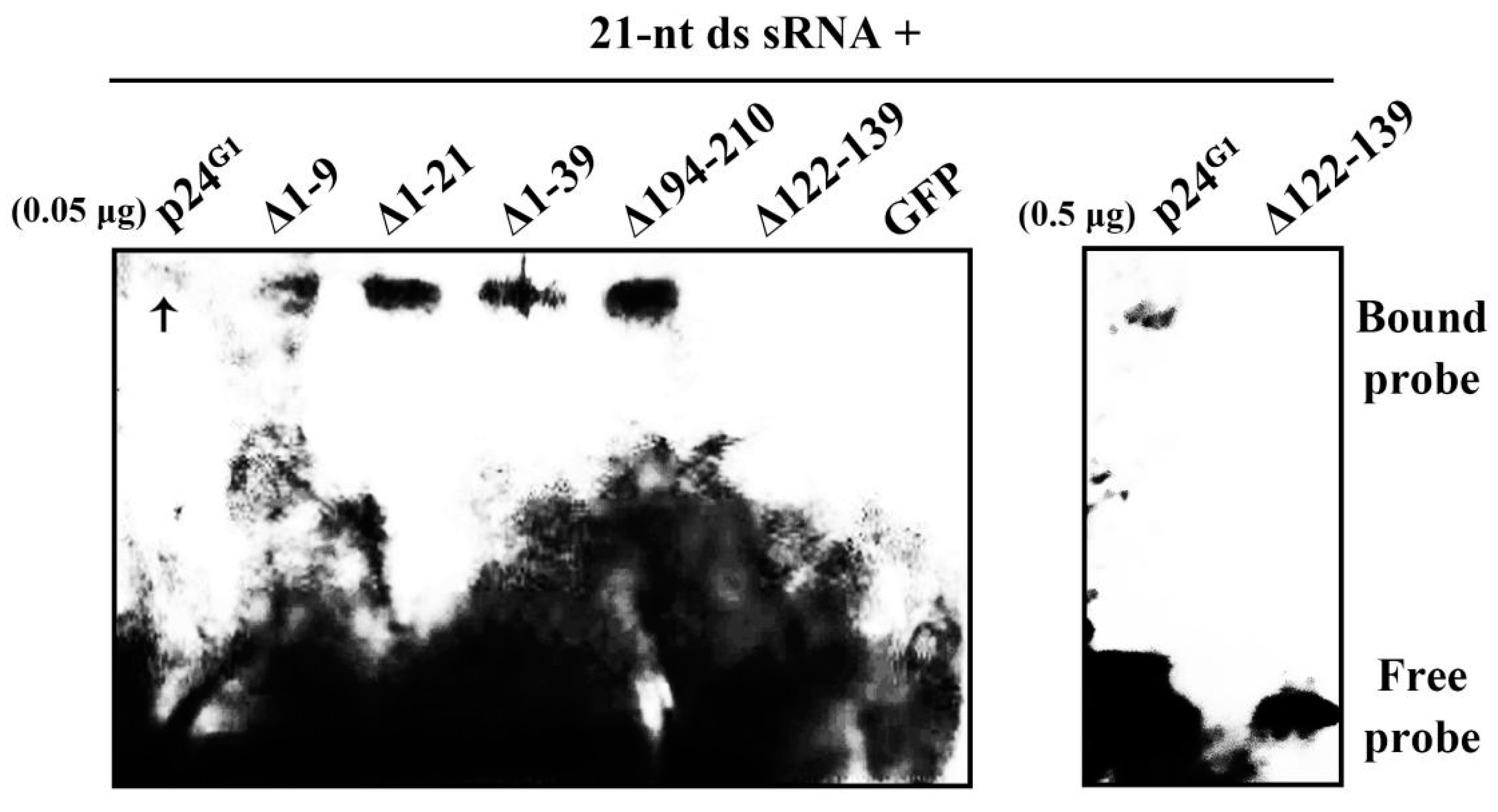
3.5. p24G1 Is Able to Bind ds siRNA
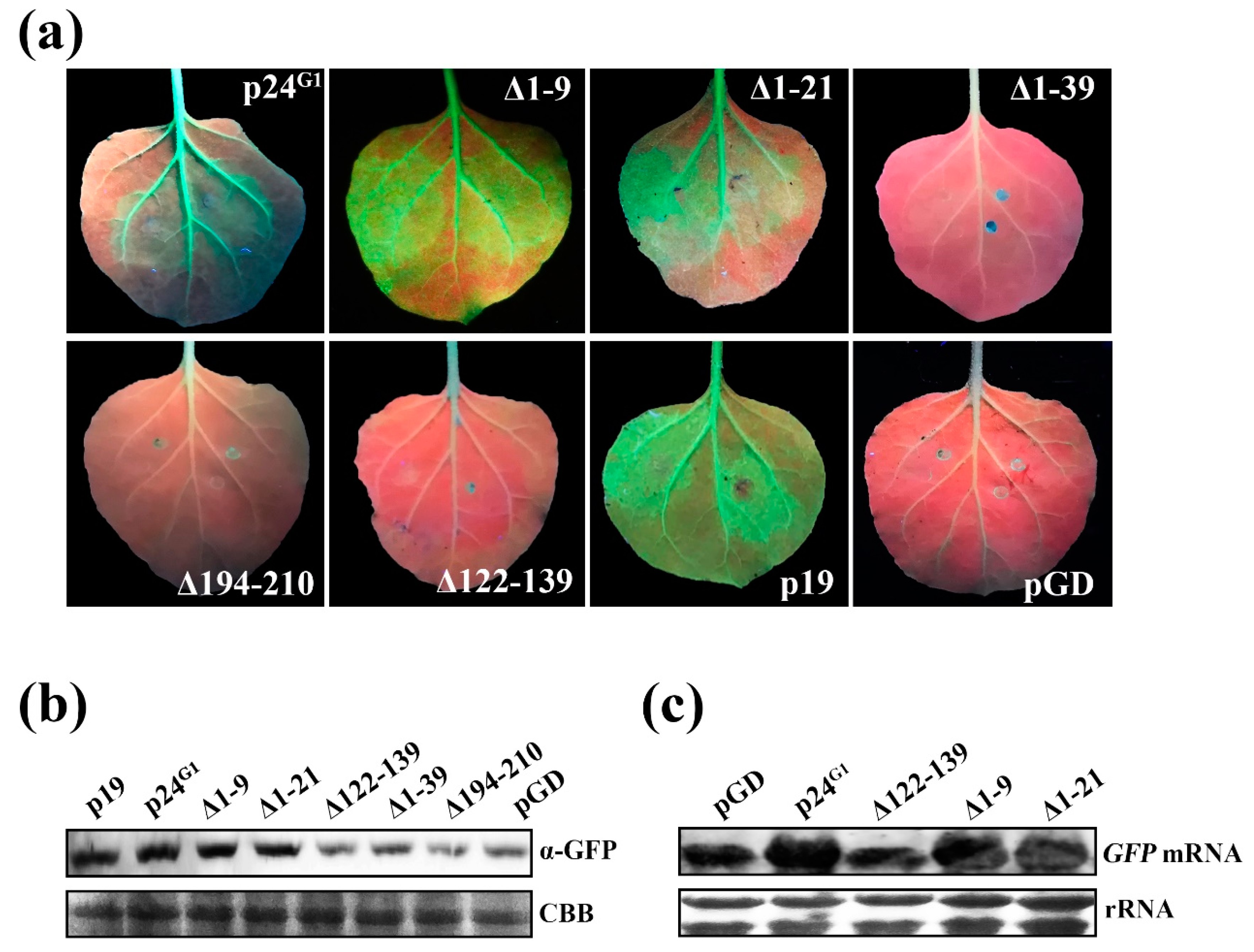
3.6. Monomeric p24G1 Can Suppress RNA Silencing, and siRNA Binding Is Insufficient for Its RSS Activity
3.7. Pathogenic Activity of p24G1 Requires Both Its RSS Activity and Dimerization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, Z.; Ebright, Y.W.; Yu, B.; Chen, X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006, 34, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.; Zhu, J.K. A viral suppressor protein inhibits host RNA silencing by hooking up with Argonautes. Genes Dev. 2010, 24, 853–856. [Google Scholar] [CrossRef] [Green Version]
- Goto, K.; Kobori, T.; Kosaka, Y.; Natsuaki, T.; Masuta, C. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007, 48, 1050–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yuan, Y.R.; Pei, Y.; Lin, S.S.; Tuschl, T.; Patel, D.J.; Chua, N.H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef] [Green Version]
- Hamera, S.; Song, X.; Su, L.; Chen, X.; Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 2012, 69, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; Lopez-Moya, J.J.; Burgyán, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, K.I.; Eskelin, K.; Bašić, M.; De, S.; Lõhmus, A.; Varjosalo, M.; Mäkinen, K. Molecular insights into the function of the viral RNA silencing suppressor HCPro. Plant J. 2016, 85, 30–45. [Google Scholar] [CrossRef]
- Vargason, J.M.; Szittya, G.; Burgyán, J.; Hall, T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 2003, 115, 799–811. [Google Scholar] [CrossRef] [Green Version]
- Ye, K.; Malinina, L.; Patel, D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 2003, 18, 874–878. [Google Scholar] [CrossRef] [Green Version]
- Bragg, J.N.; Jackson, A.O. The C-terminal region of the Barley stripe mosaic virus gamma protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 2004, 5, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Yang, J.; Lin, C.; Yuan, Y.A. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 2008, 9, 754–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, A.; Zhao, Z.; Chen, W.; Zhang, H.; Liao, Q.; Chen, J.; Du, Z. Self-interaction of the cucumber mosaic virus 2b protein plays a vital role in the suppression of RNA silencing and the induction of viral symptoms. Mol. Plant Pathol. 2013, 14, 803–812. [Google Scholar] [CrossRef]
- Yang, X.; Baliji, S.; Buchmann, R.C.; Wang, H.; Lindbo, J.A.; Sunter, G.; Bisaro, D.M. Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 2007, 81, 11972–11981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.; Van Wezel, R.; Stanley, J.; Hong, Y. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 2003, 77, 7026–7033. [Google Scholar] [CrossRef] [Green Version]
- Haas, G.; Azevedo, J.; Moissiard, G.; Geldreich, A.; Himber, C.; Bureau, M.; Fukuhara, T.; Keller, M.; Voinnet, O. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 2008, 27, 2102–2112. [Google Scholar] [CrossRef] [Green Version]
- González, I.; Rakitina, D.; Semashko, M.; Taliansky, M.; Praveen, S.; Palukaitis, P.; Carr, J.P.; Kalinina, N.; Canto, T. RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA 2012, 18, 771–782. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Zhao, X.; Jiang, L.; Yang, X.; Chen, Y.; Song, X. p15 encoded by GVX is a pathogenicity factor and RNA silencing suppressor. J. Gen. Virol. 2018, 99, 1515–1521. [Google Scholar] [CrossRef]
- Lukhovitskaya, N.I.; Vetukuri, R.R.; Sama, I.; Thaduri, S.; Solovyev, A.G.; Savenkov, E.I. A viral transcription factor exhibits antiviral RNA silencing suppression activity independent of its nuclear localization. J. Gen. Virol. 2014, 95, 2831–2837. [Google Scholar] [CrossRef] [Green Version]
- Scholthof, H.B.; Scholthof, K.B.G.; Jackson, A.O. Identification of Tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a Potato virus X vector. Plant Cell 1995, 7, 1117–1157. [Google Scholar]
- Ruiz-Ruiz, S.; Soler, N.; Sánchez-Navarro, J.; Fagoaga, C.; López, C.; Navarro, L.; Moreno, P.; Peña, L.; Flores, R. Citrus tristeza virus p23: Determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis. Mol. Plant Microbe Interact. 2013, 26, 306–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, J.A.; Pallás, V. Viral factors involved in plant pathogenesis. Curr. Opin. Virol. 2015, 11, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Naidu, R.A.; Maree, H.J.; Burger, J.T. Grapevine leafroll disease and associated viruses: A unique pathosystem. Annu. Rev. Phytopathol. 2015, 53, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, C.F.; Rezaian, M.A. Nucleotide sequence and organization of ten open reading frames in the genome of grapevine leafroll-associated virus 1 and identification of three subgenomic RNAs. J. Gen. Virol. 2000, 81, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Folimonov, A.; Shintaku, M.; Li, W.X.; Falk, B.W.; Dawson, W.O.; Ding, S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 2004, 101, 15742–15747. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.C.; Kasschau, K.D.; Prokhnevsky, A.I.; Gopinath, K.; Pogue, G.P.; Carrington, J.C.; Dolja, V.V. Suppressor of RNA silencing encoded by Beet yellows virus. Virology 2003, 306, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Li, M.J.; Zhang, J.; Feng, M.; Wang, X.Y.; Luo, C.; Wang, Q.; Cheng, Y.Q. Characterization of silencing suppressor p24 of Grapevine leafroll-associated virus 2. Mol. Plant Pathol. 2018, 19, 355–368. [Google Scholar] [CrossRef] [Green Version]
- Gouveia, P.; Nolasco, G. The p19.7 RNA silencing suppressor from Grapevine leafroll-associated virus 3 shows different levels of activity across phylogenetic groups. Virus Genes 2012, 45, 333–339. [Google Scholar] [CrossRef]
- Dey, K.K.; Borth, W.B.; Melzer, M.J.; Wang, M.L.; Hu, J.S. Analysis of pineapple mealybug wilt associated virus-1 and -2 for potential RNA silencing suppressors and pathogenicity factors. Viruses 2015, 7, 969–995. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.; Chaban, C.; Schütze, K.; Batistic, O.; Weckermann, K.; Näke, C.; Blazevic, D.; Grefen, C.; Schumacher, K.; Oecking, C.; et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004, 40, 428–438. [Google Scholar] [CrossRef]
- Goodin, M.M.; Dietzgen, R.G.; Schichnes, D.; Ruzin, S.; Jackson, A.O. pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002, 31, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Kavanagh, T.; Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 1992, 2, 549–557. [Google Scholar] [PubMed]
- Chen, L.; Yan, Z.; Xia, Z.; Cheng, Y.; Jiao, Z.; Sun, B.; Zhou, T.; Fan, Z. A violaxanthin deepoxidase interacts with a viral suppressor of RNA silencing to inhibit virus amplification. Plant Physiol. 2017, 175, 1774–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, C.G.; Fang, Y.Y.; Zhou, B.J.; Zhao, J.H.; Hou, W.N.; Zhu, H.; Ding, S.W.; Guo, H.S. Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 2012, 24, 259–274. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Voinnet, O.; Baulcombe, D. Systemic silencing in gene silencing. Nature 1997, 389, 553. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef]
- Hwang, I.S.; Choi, D.S.; Kim, N.H.; Kim, D.S.; Hwang, B.K. Pathogenesis-related protein 4b interacts with leucine-rich repeat protein1 to suppress PR4b-triggered cell death and defense response in pepper. Plant J. 2014, 77, 521–533. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.; Lange, C.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Mérai, Z.; Kerényi, Z.; Molnár, A.; Barta, E.; Válóczi, A.; Bisztray, G.; Havelda, Z.; Burgyán, J.; Silhavy, D. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005, 79, 7217–7226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuellar, W.J.; Kreuze, J.F.; Rajamäki, M.L.; Cruzado, K.R.; Untiveros, M.; Valkonen, J.P.T. Elimination of antiviral defense by a viral RNase III. Proc. Natl. Acad. Sci. USA 2009, 106, 10354–10358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tzfira, T.; Gaba, V.; Citovsky, V.; Palukaitis, P.; GalOn, A. Functional analysis of the Cucumber mosaic virus 2b protein: Pathogenicity and nuclear localization. J. Gen. Virol. 2004, 85, 3135–3147. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Qu, F.; Morris, T.J. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 2000, 12, 1917–1926. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; DelGrosso, L.; Yigit, E.; Dempsey, D.A.; Klessig, D.F.; Wobbe, K.K. The amino terminus of the coat protein of Turnip crinkle virus is the AVR factor recognized by resistant arabidopsis. Mol. Plant Microbe Interact. 2000, 13, 1015–1018. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, X.; Wu, J.; Briddon, R.W.; Zhou, X. βC1 encoded by tomato yellow leaf curl China betasatellite forms multimeric complexes in vitro and in vivo. Virology 2011, 409, 156–162. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.-W.; Liu, Q.; Zeng, Q.; Huang, W.-T.; Wang, Q.; Cheng, Y.-Q. p24G1 Encoded by Grapevine Leafroll-Associated Virus 1 Suppresses RNA Silencing and Elicits Hypersensitive Response-Like Necrosis in Nicotiana Species. Viruses 2020, 12, 1111. https://doi.org/10.3390/v12101111
Zhang C-W, Liu Q, Zeng Q, Huang W-T, Wang Q, Cheng Y-Q. p24G1 Encoded by Grapevine Leafroll-Associated Virus 1 Suppresses RNA Silencing and Elicits Hypersensitive Response-Like Necrosis in Nicotiana Species. Viruses. 2020; 12(10):1111. https://doi.org/10.3390/v12101111
Chicago/Turabian StyleZhang, Chen-Wei, Qing Liu, Qi Zeng, Wen-Ting Huang, Qi Wang, and Yu-Qin Cheng. 2020. "p24G1 Encoded by Grapevine Leafroll-Associated Virus 1 Suppresses RNA Silencing and Elicits Hypersensitive Response-Like Necrosis in Nicotiana Species" Viruses 12, no. 10: 1111. https://doi.org/10.3390/v12101111




