Emergence and Full Genome Analysis of Tomato Torrado Virus in South Africa
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA Extraction
2.2. Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
2.3. Cloning and Sequencing
2.4. Genome Analysis
3. Results
3.1. Survey Analysis
3.2. Virus Detection
3.3. Sequence Analysis
3.4. RNA-1
3.5. RNA-2
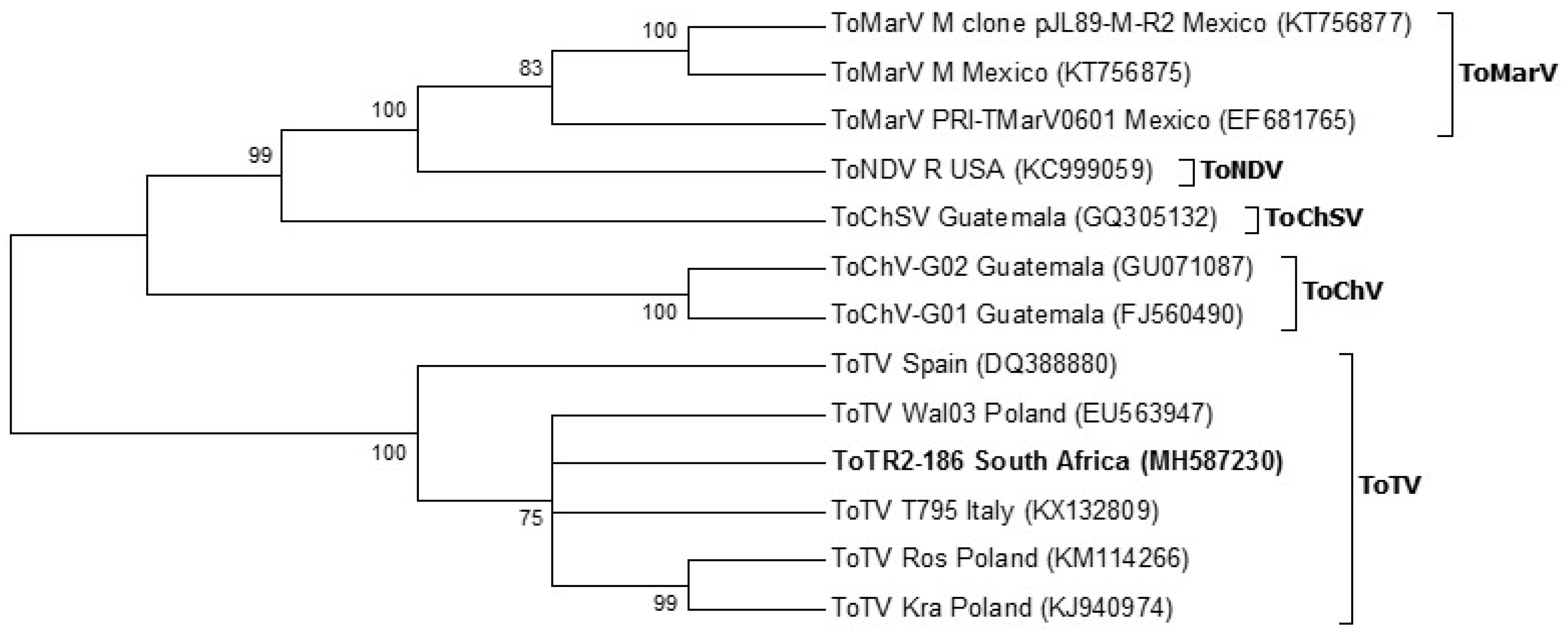
3.6. Phylogeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Verbeek, M.; Dullemans, A.M.; van den Heuvel, J.F.J.M.; Maris, P.C.; van der Vlugt, R.A.A. Identification and characterization of tomato torrado virus, a new plant picorna-like virus from tomato. Arch. Virol. 2007, 152, 881–890. [Google Scholar] [PubMed] [Green Version]
- Sanfacon, H.; Iwanami, T.; Karasev, A.V.; van der Vlugt, R.; Wellink, J.; Wetzel, T.; Yoshikawa, N. “Family Secoviridae”. In Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses; King, A., Adams, M.J., Carstens, E.B., Lefkowitz, E., Eds.; Elsevier-Academic Press: Amsterdam, The Netherlands, 2011; pp. 881–899. [Google Scholar]
- Le Gall, O.; Christian, P.; Fauquet, C.M.; King, A.M.; Knowles, N.J.; Nakashima, N.; Stanway, G.; Gorbalenya, A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008, 153, 715–727. [Google Scholar]
- Chandrasekar, V.; Johnson, J.E. The structure of tobacco ringspot virus: A link in the evolution of icosahedral capsids in the picornavirus superfamily. Structure 1998, 6, 157–171. [Google Scholar] [PubMed] [Green Version]
- Verbeek, M.; van Bekkum, P.; Dullemans, A.; van der Vlugt, R.A.A. Semi-persistent and stylet-borne manner by three whitefly vectors. Virus Res. 2014, 186. [Google Scholar]
- Wintermantel, W.M.; Hladky, L.L.; Cortez, A.A. Genome sequence, host range, and whitefly transmission of the torradovirus Tomato necrotic dwarf virus. Acta Hortic. 2018, 1207, 295–302. [Google Scholar]
- European and Mediterranean Plant Protection Organization (EPPO). 2009. Tomato Torrado Virus. Available online: https://www.eppo.int/QUARANTINE/Alert_List/.../Tomato_torrado_virus.docx./ (accessed on 22 January 2017).
- Jones, R.A.C. Future scenarios for plant virus pathogens as climate change progresses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2016; Volume 95, pp. 87–147. [Google Scholar]
- Moodley, V.; Gubba, A.; Mafongoya, P.L. A survey of whitefly-transmitted viruses on tomato crops in South Africa. Crop. Prot. 2019, 123, 21–29. [Google Scholar]
- Alfaro-Fernández, A.; Córdoba-Sellés, C.; Cebrián, M.C.; Sánchez-Navarro, J.A.; Espino, A.; Martín, R.; Jordá, C. First report of tomato torrado virus in tomato in the Canary Islands, Spain. Plant. Dis. 2007, 91, 1060. [Google Scholar]
- Pospieszny, H.; Borodynko, N.; Obrepalska-Steplowska, A.; Hasiow, B. The first report of tomato torrado virus in Poland. Plant. Dis. 2007, 91, 364. [Google Scholar]
- Alfaro-Fernández, A.; Bese, G.; Córdoba-Sellés, C.; Cebrián, M.C.; Herrera-Vásquez, J.A.; Forray, A.; Jordá, C. First report of Tomato torrado virus infecting tomato in Hungary. Plant. Dis. 2009, 93, 554. [Google Scholar]
- Herrera-Vasquez, J.A.; Alfaro-Fernández, A.; Cordoba-Selles, M.C.; Cebrian, M.C.; Font, M.I.J.C. First report of tomato torrado virus infecting tomato in single and mixed infections with cucumber mosaic virus in Panama. Plant. Dis. 2009, 93, 198. [Google Scholar]
- Verdin, E.; Gognalons, P.; Wipf-Scheibel, C.; Bornard, I.; Ridray, G.; Schoen, L.; Lecoq, H. First report of tomato torrado virus in tomato crops in France. Plant. Dis. 2009, 93, 1352. [Google Scholar] [CrossRef]
- Davino, S.; Bivona, L.; Iacono, G.; Davino, M. First report of Tomato torrado virus infecting tomato in Italy. Plant. Dis. 2010, 94, 1172. [Google Scholar] [CrossRef] [PubMed]
- Gambley, C.F.; Thomas, J.E.; Persley, D.M.; Hall, B.H. First report of Tomato torrado virus on tomato from Australia. Plant. Dis. 2010, 94, 486. [Google Scholar] [PubMed]
- Verbeek, M.; Dullemans, A. First Report of Tomato torrado virus infecting tomato in Colombia. Plant. Dis. 2012, 96, 592. [Google Scholar] [CrossRef] [PubMed]
- European and Mediterranean Plant Protection Organization (EPPO). 2018. Available online: https://gd.eppo.int/taxon/TOTV00/distribution/MA/ (accessed on 15 April 2018).
- Moodley, V.; Gubba, A.; Mafongoya, P.L. First Report of Tomato torrado virus on tomato (Solanum lycopersicum) in South Africa. Plant. Dis. 2016, 100, 231. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Budziszewska, M.; Obrepalska-Steplowska, A.; Wieczorek, P.; Pospieszny, H. The nucleotide sequence of a Polish isolate of Tomato torrado virus. Virus Genes 2008, 37, 400. [Google Scholar] [CrossRef]
- Mushegian, A.R. The putative movement domain encoded by nepovirus RNA-2 is conserved in all sequenced nepoviruses. Arch. Virol. 1994, 135, 437–441. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Gomez, P.; Sempere, R.N.; Amari, K.; Gomez-Aix, C.; Aranda, M.A. Epidemics of Tomato torrado virus, Pepino mosaic virus and Tomato chlorosis virus in tomato crops: Do mixed infections contribute to torrado disease epidemiology? Ann. Appl. Biol. 2010, 156, 401–410. [Google Scholar] [CrossRef]
- Alfaro-Fernández, A.; Córdoba-Sellés, M.C.; Juárez, M.; Herrera-Vásquez, J.A.; Sánchez-Navarro, J.A.; Cebrián, M.C.; Font, M.I.; Jordá, C. Occurrence and geographical distribution of the ‘Torrado’ disease in Spain. J. Phytopathol. 2010, 158, 457–469. [Google Scholar] [CrossRef]
- Pospieszny, H.; Budziszewska, M.; Hasiów-Jaroszewska, B.; Obrępalska-Stęplowska, A.; Borodynko, N. Biological and molecular characterization of Polish isolates of Tomato torrado virus. J. Phytopathol. 2010, 158, 56–62. [Google Scholar] [CrossRef]
- Pospieszny, H.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B.; Rymelska, N.; Elena, S.F. Transmission rate of two Polish Tomato torrado virus isolates through tomato seeds. J. Gen. Plant. Pathol. 2019, 85, 109. [Google Scholar] [CrossRef] [Green Version]
- Van der Vlugt, R.A.A.; Verbeek, M.; Dullemans, A.M.; Wintermantel, W.M.; Cuellar, W.J.; Fox, A.; Thompson, J.R. Torradoviruses. Annu. Rev. Phytopathol. 2015, 53, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Pospieszny, H.; Borodynko, N.; Hasiow-Jaroszewska, B.; Rymelska, N. Seed transmission of Tomato torradovirus in tomato. In Proceedings of the 12th International Plant Virus Epidemiology Symposium, Arusha, Tanzania, 28 January–1 February 2013. [Google Scholar]
- Pospieszny, H.; Borodynko, N. Seed transmission of Tomato torrado virus in pepper (Capsicum annuum). In Proceedings of the 16th International Congress for Molecular Plant-Microbe Interactions, Rhodes, Greece, 6–10 July 2014; p. 168. [Google Scholar]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Torradovirus Isolate (RNA 1) | 5′UTR | Polyprotein | ORF 1 Polyprotein | 3′UTR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protease co. | Helicase | Protease | RdRP | |||||||||
| nt (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | |
| ToT-186 (MH587229) South Africa | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| ToTV T795 (KX132808) Italy | 99.06 | 99.05 | 99.72 | 99.71 | 100 | 98.97 | 100 | 99.33 | 100 | 98.94 | 100 | 97.82 |
| ToTV Ros (KM091449) Poland | 98.13 | 99.07 | 99.30 | 99.42 | 99.56 | 99.31 | 100 | 100 | 100 | 99.07 | 100 | 97.36 |
| ToTV Wal03 (EU563948) Poland | 100 | 98.96 | 99.53 | 99.71 | 100 | 98.97 | 100 | 99.66 | 100 | 98.94 | 99.60 | 99.50 |
| ToTV Kra (KJ940975) Poland | 100 | 99.10 | 99.49 | 99.57 | 100 | 98.97 | 100 | 99 | 100 | 99.07 | 100 | 99.67 |
| ToTV (DQ388879) Spain | 98.13 | 98.98 | 99.58 | 99.71 | 100 | 98.63 | 100 | 99.66 | 100 | 99.80 | 100 | 98.76 |
| ToChV-G01 (FJ560489) Guatemala | 72.89 | 38.20 | 30.56 | N/A | N/A | 24.82 | 14.43 | N/A | N/A | 72.02 | 87.95 | 61.19 |
| ToChSV (GQ305131) Guatemala | 71.02 | 38.01 | 31.86 | N/A | N/A | 30.30 | 21.21 | 30.34 | 14.92 | 24.20 | 13.09 | 62.05 |
| ToMarV PRI (EF681764) Mexico | 63.55 | 26.34 | 13.38 | N/A | N/A | 79.59 | 98.96 | N/A | N/A | 26.23 | 13.25 | N/A |
| ToMarV pJL89 (KT756876) Mexico | 66.35 | 26.53 | 13.42 | N/A | N/A | 79.59 | 98.96 | N/A | N/A | 25.97 | 13.25 | 61.24 |
| ToMarV M (KT756874) Mexico | 66.35 | 26.56 | 13.42 | N/A | N/A | 79.59 | 98.96 | N/A | N/A | 26.10 | 13.25 | 61.40 |
| ToNDV R (KC999058) USA | 66.35 | 48.56 | 49.83 | N/A | N/A | 78.91 | 97.93 | N/A | N/A | N/A | N/A | 62.24 |
| Torradovirus Isolate (RNA-2) | ORF-2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′UTR | ORF 1 | MP | VP 35 | VP 26 | VP 23 | 3′UTR | ||||||
| nt (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | aa (%) | nt (%) | |
| ToT-186 (MH587230) South Africa | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| ToTV T795 (KX132809) Italy | 97.34 | 99.47 | 98.94 | 99.08 | 100 | 98.78 | 100 | 99.43 | 100 | 99.38 | 99.53 | 99.53 |
| ToTV Ros (KM114266) Poland | 97.87 | 98.95 | 98.94 | 98.93 | 100 | 98.64 | 99.18 | 99.01 | 99.57 | 99.53 | 99.53 | 99.53 |
| ToTV Wal03 (EU563947) Poland | 97.87 | 99.3 | 99.47 | 98.48 | 100 | 99.05 | 100 | 99.29 | 100 | 99.07 | 100 | 99.59 |
| ToTV Kra (KJ940974) Poland | 97.34 | 99.47 | 99.47 | 98.63 | 99.08 | 98.5 | 99.59 | 99.15 | 100 | 99.53 | 99.53 | 99.53 |
| ToTV (DQ388880) Spain | 97.34 | 98.6 | 98.94 | 98.93 | 100 | 98.91 | 100 | 99.01 | 100 | 98.92 | 99.53 | 99.46 |
| ToChV-G01 (FJ560490) Guatemala | 46.27 | 57.41 | 70 | 70.06 | 80.36 | 63.27 | 73.87 | 73.55 | 87.28 | 65.74 | 81.94 | 31.43 |
| ToChV-G02 (GU071087) Guatemala | 46.27 | 60.73 | 71.05 | 71.27 | 80.36 | 63.55 | 73.87 | 73.69 | 86.86 | 65.59 | 80.55 | 31.43 |
| ToChSV (GQ305132) Guatemala | 40.42 | 63.52 | 73.68 | 70.36 | 80.82 | 64.9 | 74.69 | 74.68 | 91.52 | 64.36 | 81.94 | 49.23 |
| PRI-TMarV0601 (EF681765) Mexico | 36.7 | 60.73 | 72.1 | 70.36 | 80.82 | 64.22 | 76.32 | 73.41 | 89.83 | 65.74 | 79.62 | 45.88 |
| ToMarV pJL89 (KT756877) Mexico | 36.17 | 60.38 | 73.68 | 70.06 | 81.27 | 66.66 | 75.51 | 73.41 | 89.4 | 66.05 | 80.55 | 46.62 |
| ToMarV M (KT756875) Mexico | 36.17 | 60.55 | 73.68 | 69.9 | 81.27 | 66.53 | 75.51 | 73.55 | 89.4 | 65.89 | 80.55 | 46.62 |
| ToNDV R (KC999059) USA | 35.63 | 61.6 | 70.52 | 70.06 | 81.27 | 64.9 | 74.28 | 72.15 | 91.1 | 66.35 | 80.55 | 46.48 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moodley, V.; Gubba, A.; Mafongoya, P.L. Emergence and Full Genome Analysis of Tomato Torrado Virus in South Africa. Viruses 2020, 12, 1167. https://doi.org/10.3390/v12101167
Moodley V, Gubba A, Mafongoya PL. Emergence and Full Genome Analysis of Tomato Torrado Virus in South Africa. Viruses. 2020; 12(10):1167. https://doi.org/10.3390/v12101167
Chicago/Turabian StyleMoodley, Vaneson, Augustine Gubba, and Paramu L. Mafongoya. 2020. "Emergence and Full Genome Analysis of Tomato Torrado Virus in South Africa" Viruses 12, no. 10: 1167. https://doi.org/10.3390/v12101167
APA StyleMoodley, V., Gubba, A., & Mafongoya, P. L. (2020). Emergence and Full Genome Analysis of Tomato Torrado Virus in South Africa. Viruses, 12(10), 1167. https://doi.org/10.3390/v12101167







