Can Measurements of Inflammatory Biomarkers Be Used to Spot Respiratory Viral Infections?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort and Parameters/Biomarkers Measured
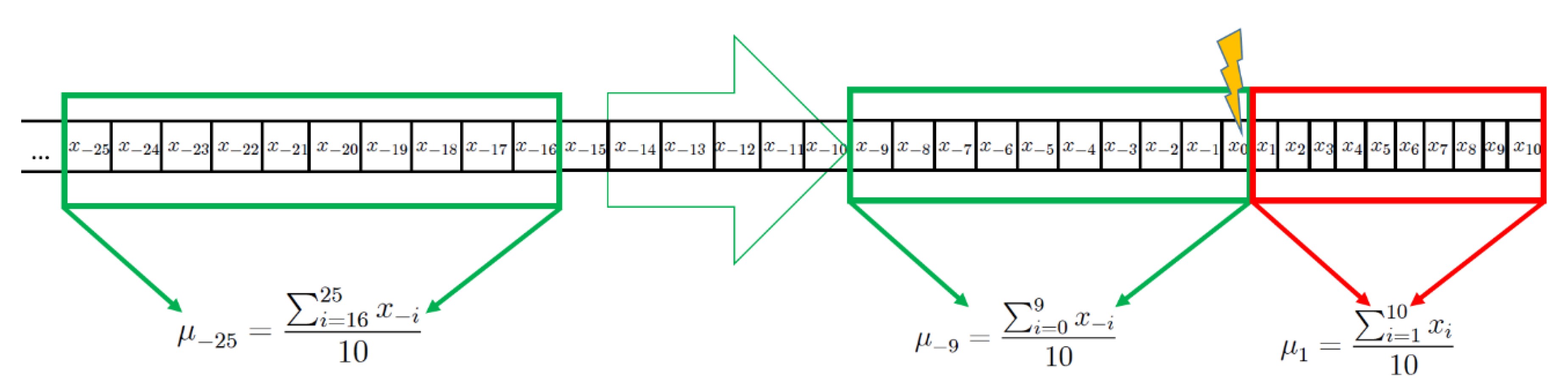
2.2. Computational Methods and Statistical Analysis
3. Results
4. Discussion
4.1. Main Findings and Interpretations
4.2. Limitations of This Study
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mahony, J. Detection of Respiratory Viruses by Molecular Methods. Clin. Microbiol. Rev. 2008, 21, 716–747. [Google Scholar] [CrossRef] [Green Version]
- Barenfanger, J.; Drake, C.; Leon, N.; Mueller, T.; Troutt, T. Clinical and Financial Benefits of Rapid Detection of Respiratory Viruses: An Outcomes Study. J. Clin. Microbiol. 2000, 38, 2824–2828. [Google Scholar] [CrossRef] [Green Version]
- Nicolini, A.; Ferrera, L.; Rao, F.; Senarega, R.; Ferrari-Bravo, M. Chest radiological findings of influenza A H1N1 pneumonia. Rev. Port. de Pneumol. 2012, 18, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peaper, D.R.; Landry, M.L. Laboratory diagnosis of viral infection. Neurogenetics Part I 2014, 123, 123–147. [Google Scholar] [CrossRef]
- Bray, M.; Lawler, J.; Paragas, J.; Jahrling, P.B.; Mollura, D.J. Molecular Imaging of Influenza and Other Emerging Respiratory Viral Infections. J. Infect. Dis. 2011, 203, 1348–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, F.; He, J.; Loo, J.F.C.; Yao, J.; Shi, L.; Liu, C.; Zhao, C.; Xie, W.; Shao, Y.; Kong, S.K.; et al. Identification of microRNAs in Throat Swab as the Biomarkers for Diagnosis of Influenza. Int. J. Med Sci. 2016, 13, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Abels, S.; Nadal, D.; Stroehle, A.; Bossart, W. Reliable Detection of Respiratory Syncytial Virus Infection in Children for Adequate Hospital Infection Control Management. J. Clin. Microbiol. 2001, 39, 3135–3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andeweg, A.C.; Bestebroer, T.M.; Huybreghs, M.; Kimman, T.G.; De Jong, J.C. Improved Detection of Rhinoviruses in Clinical Samples by Using a Newly Developed Nested Reverse Transcription-PCR Assay. J. Clin. Microbiol. 1999, 37, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woloshin, S.; Patel, N.; Kesselheim, A.S. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N. Engl. J. Med. 2020, 383, e38. [Google Scholar] [CrossRef]
- Hui, R.K.H.; Zeng, F.; Chan, C.M.N.; Yuen, K.-Y.; Peiris, J.S.M.; Leung, F.C.C.; Giraldo, M.; Portela, R.W.D.; Snege, M.; Leser, P.G.; et al. Reverse Transcriptase PCR Diagnostic Assay for the Coronavirus Associated with Severe Acute Respiratory Syndrome. J. Clin. Microbiol. 2004, 42, 1400–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yam, W.C.; Chan, K.H.; Poon, L.L.M.; Guan, Y.; Yuen, K.Y.; Seto, W.H.; Peiris, J.S.M. Evaluation of Reverse Transcription-PCR Assays for Rapid Diagnosis of Severe Acute Respiratory Syndrome Associated with a Novel Coronavirus. J. Clin. Microbiol. 2003, 41, 4521–4524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Dunbar, S.; Tang, Y.-W. Laboratory Diagnosis of Respiratory Tract Infections in Children—The State of the Art. Front. Microbiol. 2018, 9, 2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez, Y.; González, L.; Noguera, L.; González, P.A.; Riedel, C.A.; Bertrand, P.; Bueno, S.M. Cytokines in the Respiratory Airway as Biomarkers of Severity and Prognosis for Respiratory Syncytial Virus Infection: An Update. Front. Immunol. 2019, 10, 10. [Google Scholar] [CrossRef]
- Sinha, A.; Lutter, R.; Xu, B.; Dekker, T.; Dierdorp, B.; Sterk, P.J.; Frey, U.; Delgado-Eckert, E. Loss of adaptive capacity in asthmatic patients revealed by biomarker fluctuation dynamics after rhinovirus challenge. eLife 2019, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Van Der Sluijs, K.F.; Van De Pol, M.A.; Kulik, W.; Dijkhuis, A.; Smids, B.S.; Van Eijk, H.W.; Karlas, J.A.; Molenkamp, R.; Wolthers, K.C.; Johnston, S.L.; et al. Systemic tryptophan and kynurenine catabolite levels relate to severity of rhinovirus-induced asthma exacerbation: A prospective study with a parallel-group design. Thorax 2013, 68, 1122–1130. [Google Scholar] [CrossRef] [Green Version]
- Yusa, T.; Tateda, K.; Ohara, A.; Miyazaki, S. New possible biomarkers for diagnosis of infections and diagnostic distinction between bacterial and viral infections in children. J. Infect. Chemother. 2017, 23, 96–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denlinger, L.C.; Sorkness, R.L.; Lee, W.-M.; Evans, M.D.; Wolff, M.J.; Mathur, S.K.; Crisafi, G.M.; Gaworski, K.L.; Pappas, T.E.; Vrtis, R.F.; et al. Lower Airway Rhinovirus Burden and the Seasonal Risk of Asthma Exacerbation. Am. J. Respir. Crit. Care Med. 2011, 184, 1007–1014. [Google Scholar] [CrossRef] [Green Version]
- Zweig, M.H.; Campbell, G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [CrossRef]
- Šimundić, A.M. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC 2009, 19, 203–211. [Google Scholar]
- Kelly, J.T.; Busse, W.W. Host immune responses to rhinovirus: Mechanisms in asthma. J. Allergy Clin. Immunol. 2008, 122, 671–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey, R.T., Jr.; Lynfield, R.; Dwyer, D.E.; Losso, M.H.; Cozzi-Lepri, A.; Wentworth, D.; Lane, H.C.; Dewar, R.; Rupert, A.; Metcalf, J.A.; et al. The Association between Serum Biomarkers and Disease Outcome in Influenza A(H1N1)pdm09 Virus Infection: Results of Two International Observational Cohort Studies. PLOS ONE 2013, 8, e57121. [Google Scholar] [CrossRef]
- Smith, S. Digital Signal Processing: A Practical Guide for Engineers and Scientists; Elsevier: Burlinton, MA, USA, 2013. [Google Scholar]
- Bourdin, A.; Gras, D.; Vachier, I.; Chanez, P. Upper airway·1: Allergic rhinitis and asthma: United disease through epithelial cells. Thorax 2009, 64, 999–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohde, G.; Message, S.D.; Haas, J.J.; Kebadze, T.; Parker, H.; Laza-Stanca, V.; Khaitov, M.R.; Kon, O.M.; Stanciu, L.A.; Mallia, P.; et al. CXC chemokines and antimicrobial peptides in rhinovirus-induced experimental asthma exacerbations. Clin. Exp. Allergy 2014, 44, 930–939. [Google Scholar] [CrossRef] [Green Version]
- Lewis, T.C.; Henderson, T.A.; Carpenter, A.R.; Ramirez, I.A.; McHenry, C.L.; Goldsmith, A.M.; Ren, X.; Mentz, G.B.; Mukherjee, B.; Robins, T.G.; et al. Nasal cytokine responses to natural colds in asthmatic children. Clin. Exp. Allergy 2012, 42, 1734–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Demographic Features | Healthy | Asthmatic |
|---|---|---|
| Total number, n | 12 | 12 |
| Female gender, n (%) | 7 (58.3%) | 8 (66.7%) |
| Age (years), mean (SD) | 21 ± 1.5 | 22.2 ± 2.2 |
| Ethnicity (Caucasian), n (non-Caucasian, n) | 11 | 9 |
| BMI, mean (SD) * | 22.2 ± 1.6 | 22.8 ± 3.1 |
| Smoking (pack years), n | 1 (0.17 PY) | -- |
| Height (centimeters) * | 177.7 ± 8.6 | 172.5 ± 13.0 |
| Weight (KG) * | 70.4 ± 10.1 | 67.8 ± 12.4 |
| Baseline spirometry measurements | ||
| FEV1 % predicted * | 105.7 ± 11.6 | 101.0 ± 10.0 |
| FVC % predicted * | 104.2 ± 10.5 | 104.2 ± 10.2 |
| PEF % predicted * | 108.4 ± 14.0 | 104.7 ± 12.2 |
| Healthy | Asthmatics |
|---|---|
| No history of episodic chest symptoms | History of episodic chest symptoms |
| Baseline FEV1 ≥ 80 % predicted | Baseline FEV1 ≥ 70% predicted |
| AHR to methacholine (PC20) ≥ 19.6 mg/mL | AHR to methacholine (PC20) ≤ 9.8 mg/mL |
| SPT negative for all 12 common Aeroallergens | SPT positive for at least 1 out of 12 common Aeroallergens |
| Biomarker or Parameter | Sampling Frequency before Rhinovirus Challenge | Sampling Frequency after Rhinovirus Challenge |
|---|---|---|
| Lung function (FEV1, FVC, FEV1/FVC, PEF) | 2× daily | 2× daily |
| Exhaled Nitric Oxide (FeNO) | 3× weekly | 3× weekly |
| Eosinophil and neutrophil cell numbers in nasal lavage fluid | 1× weekly | 3× weekly |
| Cytokines in nasal lavage fluid | 1× weekly | 3× weekly |
| Viral Load in Nasal Lavage Fluid Measured Using RVPCR (Copies/mL) |
Seroconversion Response | |||
|---|---|---|---|---|
| Subject ID | Day 1 after Challenge | Day 3 after Challenge | Day 6 after Challenge | |
| 01A | 4040 | 95,600 | 8520 | 1 |
| 02A | 138,000 | 0 | 4970 | 0 |
| 04A | 0 | 0 | 0 | 1 |
| 05A | NA | 359,000 | 486,000 | 0 |
| 06A | 18,000 | 1,350,000 | 14,500 | 1 |
| 07A | NA | 1,580,000 | 216,000 | 1 |
| 08A | 0 | 0 | 0 | 1 |
| 09A | NA | 62,700 | 1,240,000 | 0 |
| 10A | 0 | NA | 0 | 1 |
| 11A | 3210 | 4,310,000 | 3,690,000 | 1 |
| 12A | NA | 86,900 | NA | 0 |
| 13A | NA | 0 | NA | 0 |
| 01H | NA | 29,300 | NA | 0 |
| 03H | NA | 1,180,000 | 13,000 | 0 |
| 05H | 18,300 | 9,640,000 | 13,300 | 1 |
| 06H | 0 | 32,700 | 0 | 0 |
| 07H | 0 | 0 | 0 | 1 |
| 08H | 0 | 26,900 | NA | 1 |
| 09H | 0 | 169,000 | 7,690,000 | 1 |
| 11H | 0 | 0 | 0 | 0 |
| 12H | 2560 | 15,700 | 7160 | 1 |
| 13H | 0 | 239,000 | 2,630,000 | 1 |
| 14H | 0 | 16,600,000 | 13,600,000 | 0 |
| 15H | 0 | 0 | 220,000 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinha, A.; Lutter, R.; Dekker, T.; Dierdorp, B.; J. Sterk, P.; Frey, U.; Delgado-Eckert, E. Can Measurements of Inflammatory Biomarkers Be Used to Spot Respiratory Viral Infections? Viruses 2020, 12, 1175. https://doi.org/10.3390/v12101175
Sinha A, Lutter R, Dekker T, Dierdorp B, J. Sterk P, Frey U, Delgado-Eckert E. Can Measurements of Inflammatory Biomarkers Be Used to Spot Respiratory Viral Infections? Viruses. 2020; 12(10):1175. https://doi.org/10.3390/v12101175
Chicago/Turabian StyleSinha, Anirban, René Lutter, Tamara Dekker, Barbara Dierdorp, Peter J. Sterk, Urs Frey, and Edgar Delgado-Eckert. 2020. "Can Measurements of Inflammatory Biomarkers Be Used to Spot Respiratory Viral Infections?" Viruses 12, no. 10: 1175. https://doi.org/10.3390/v12101175
APA StyleSinha, A., Lutter, R., Dekker, T., Dierdorp, B., J. Sterk, P., Frey, U., & Delgado-Eckert, E. (2020). Can Measurements of Inflammatory Biomarkers Be Used to Spot Respiratory Viral Infections? Viruses, 12(10), 1175. https://doi.org/10.3390/v12101175





