Disruption of Zika Virus xrRNA1-Dependent sfRNA1 Production Results in Tissue-Specific Attenuated Viral Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
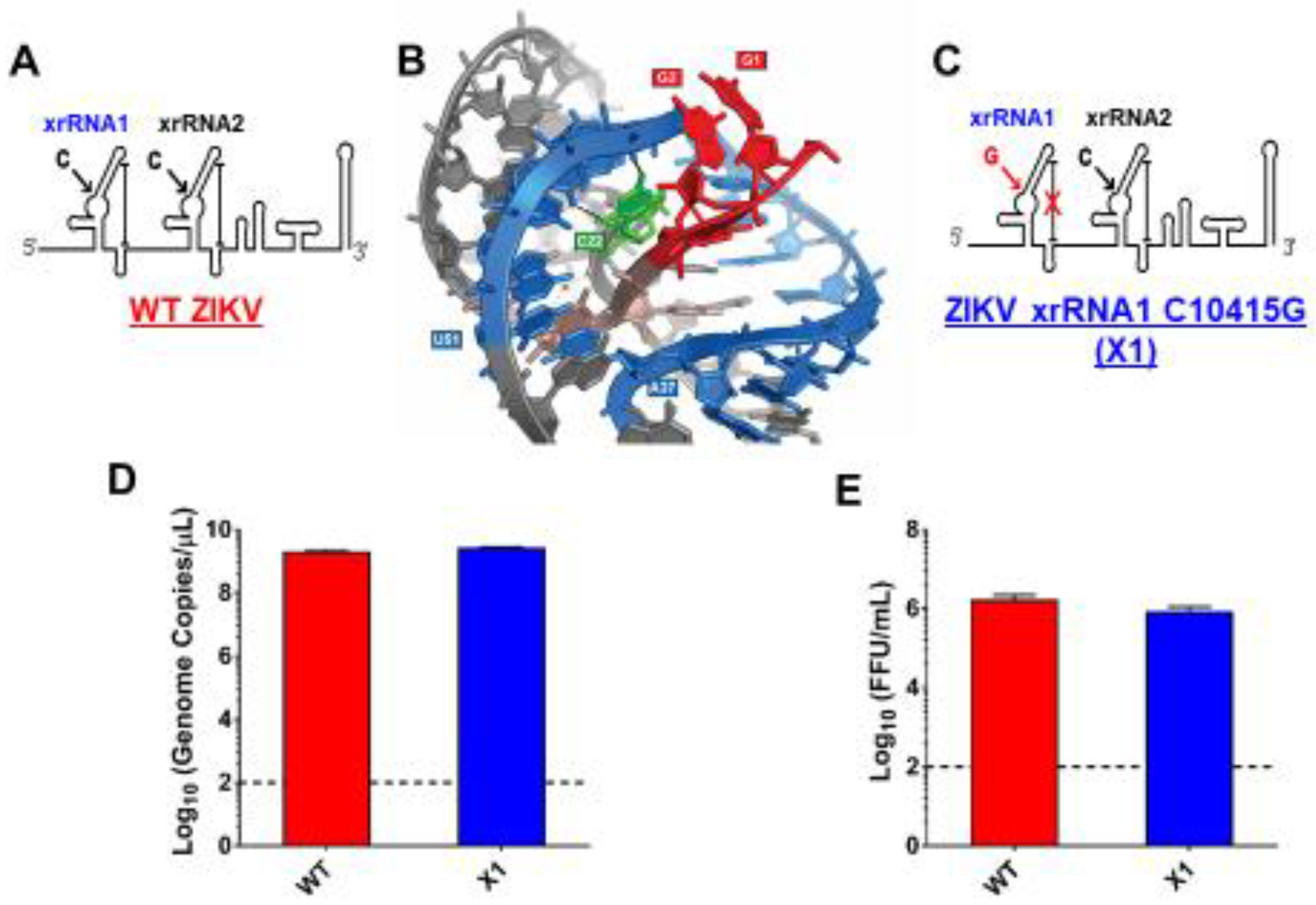
2.2. Plasmids and Generation of the X1 Mutant
2.3. Rescue and Propagation of ZIKV
2.4. Virus Quantification
2.5. Northern Blot
2.6. In Vitro Viral Growth Kinetics Comparison
2.7. Animal Studies
2.8. Characterization of Infection in Adult Ifnar1−/− Mice
2.9. Infection in Pregnant Ifnar1−/− Mice
2.10. Tissue Viral Load by RT-qPCR
2.11. Infection of Adult HuSTAT2 Mice
2.12. ZIKV-Reactive IgG ELISA
2.13. FRNT Measurement of Serum Neutralization
2.14. Statistical Analysis
3. Results
3.1. Development of an Infectious ZIKV X1 Mutant
3.2. X1 Mutation Does Not Significantly Alter Viral Growth In Vitro
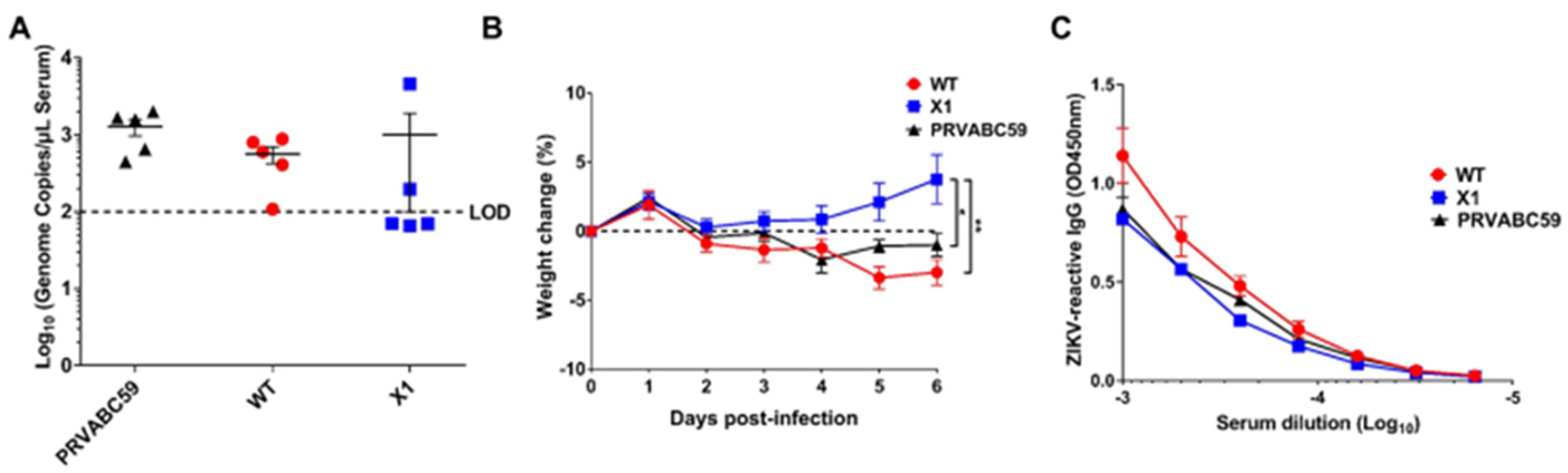
3.3. X1 ZIKV Is Attenuated in Adult Ifnar1−/− Mice and Produces Neutralizing Antibody Responses
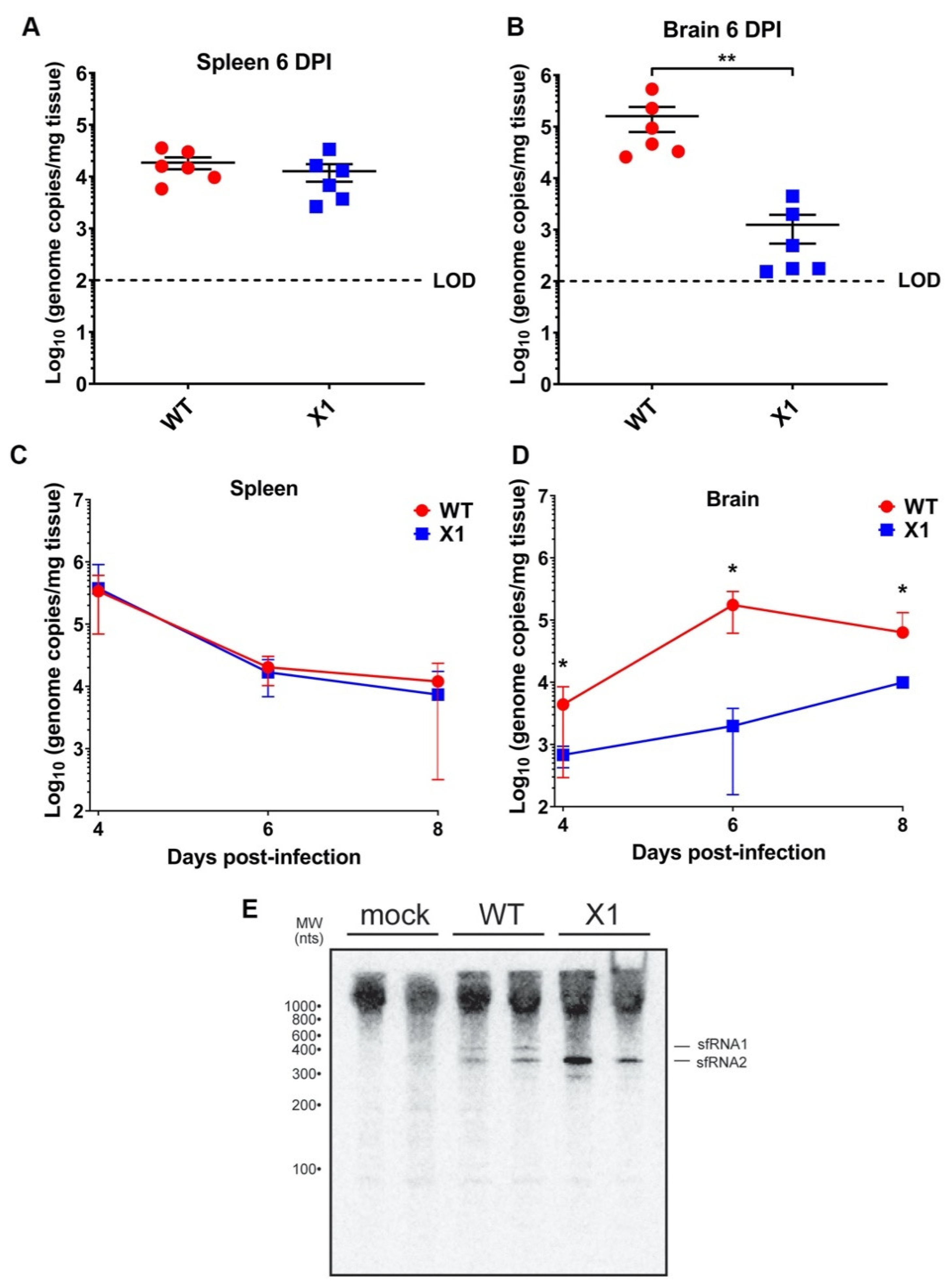
3.4. X1 Is Attenuated in the Central Nervous System (CNS) Tissue of Adult Ifnar1−/− Mice
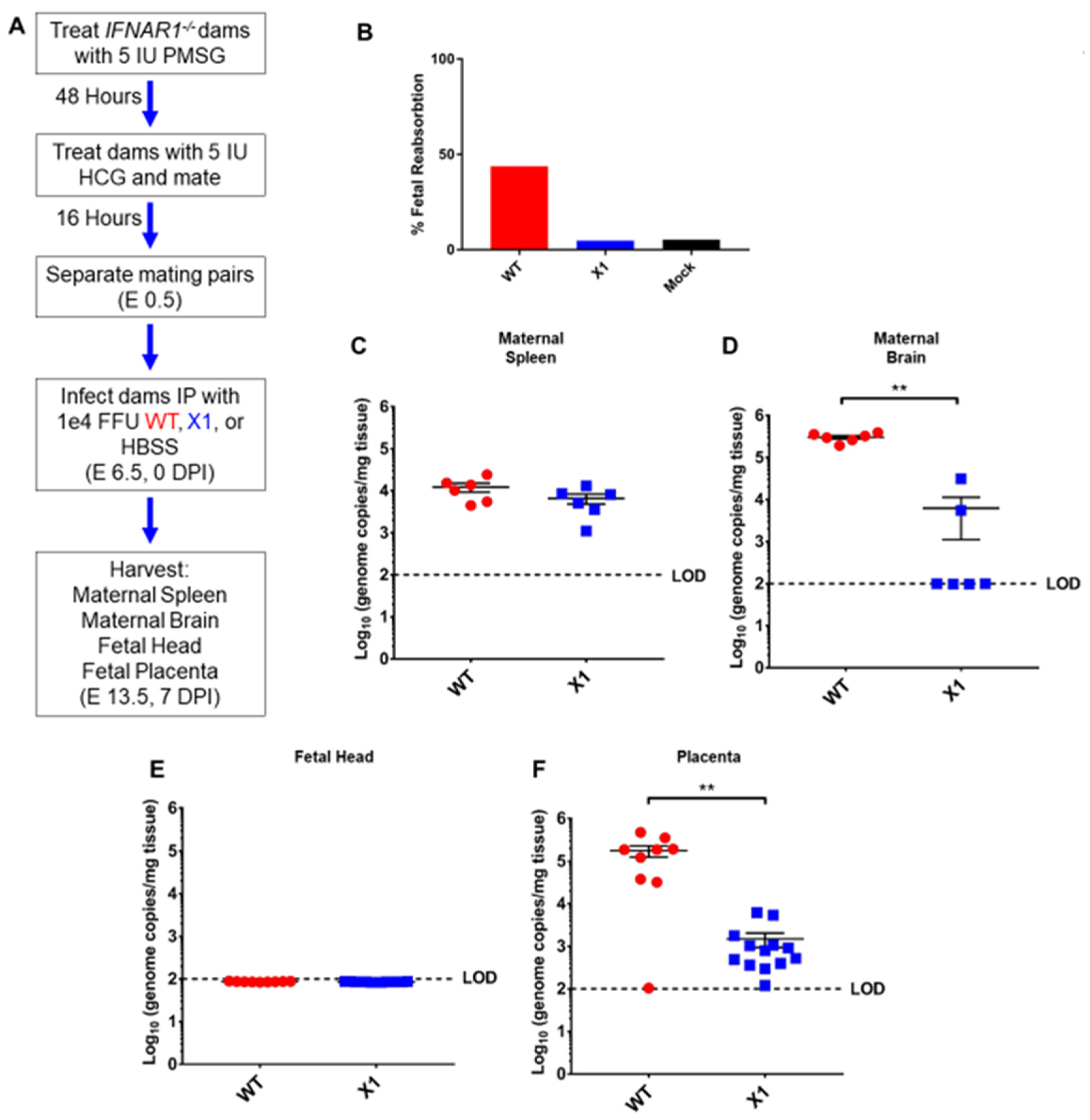
3.5. X1 ZIKV Replication Is Attenuated in the Placenta of Pregnant Ifnar1−/− Mice
3.6. Attenuated X1 ZIKV Generates a Strong Neutralizing Antibody Response in Transgenic STAT2 Mice
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Akiyama, B.M.; Laurence, H.M.; Massey, A.R.; Costantino, D.A.; Xie, X.; Yang, Y.; Shi, P.Y.; Nix, J.C.; Beckham, J.D.; Kieft, J.S. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 2016, 354, 1148–1152. [Google Scholar] [CrossRef] [Green Version]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef] [Green Version]
- Chang, R.Y.; Hsu, T.W.; Chen, Y.L.; Liu, S.F.; Tsai, Y.J.; Lin, Y.T.; Chen, Y.S.; Fan, Y.H. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet. Microbiol. 2013, 166, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Moon, S.L.; Wilusz, J.; Kieft, J.S. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife 2014, 3, e01892. [Google Scholar] [CrossRef] [PubMed]
- Goertz, G.P.; Fros, J.J.; Miesen, P.; Vogels, C.B.F.; van der Bent, M.L.; Geertsema, C.; Koenraadt, C.J.M.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes. J. Virol. 2016, 90, 10145–10159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.L.; Dodd, B.J.; Brackney, D.E.; Wilusz, C.J.; Ebel, G.D.; Wilusz, J. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology 2015, 485, 322–329. [Google Scholar] [CrossRef] [Green Version]
- Schnettler, E.; Sterken, M.G.; Leung, J.Y.; Metz, S.W.; Geertsema, C.; Goldbach, R.W.; Vlak, J.M.; Kohl, A.; Khromykh, A.A.; Pijlman, G.P. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J. Virol. 2012, 86, 13486–13500. [Google Scholar] [CrossRef] [Green Version]
- Schnettler, E.; Tykalova, H.; Watson, M.; Sharma, M.; Sterken, M.G.; Obbard, D.J.; Lewis, S.H.; McFarlane, M.; Bell-Sakyi, L.; Barry, G.; et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 2014, 42, 9436–9446. [Google Scholar] [CrossRef]
- Schuessler, A.; Funk, A.; Lazear, H.M.; Cooper, D.A.; Torres, S.; Daffis, S.; Jha, B.K.; Kumagai, Y.; Takeuchi, O.; Hertzog, P.; et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 2012, 86, 5708–5718. [Google Scholar] [CrossRef] [Green Version]
- Pallares, H.M.; Costa Navarro, G.S.; Villordo, S.M.; Merwaiss, F.; de Borba, L.; Gonzalez Lopez Ledesma, M.M.; Ojeda, D.S.; Henrion-Lacritick, A.; Morales, M.A.; Fabri, C.; et al. Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Zou, J.; Zhang, B.; Yuan, Z. Dengue virus subgenomic RNA induces apoptosis through the Bcl-2-mediated PI3k/Akt signaling pathway. Virology 2014, 448, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Weger-Lucarelli, J.; Duggal, N.K.; Bullard-Feibelman, K.; Veselinovic, M.; Romo, H.; Nguyen, C.; Ruckert, C.; Brault, A.C.; Bowen, R.A.; Stenglein, M.; et al. Development and Characterization of Recombinant Virus Generated from a New World Zika Virus Infectious Clone. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Brien, J.D.; Lazear, H.M.; Diamond, M.S. Propagation, quantification, detection, and storage of West Nile virus. Curr. Protoc. Microbiol. 2013, 31, 15D-3. [Google Scholar]
- Luo, C.; Zuniga, J.; Edison, E.; Palla, S.; Dong, W.; Parker-Thornburg, J. Superovulation strategies for 6 commonly used mouse strains. J. Am. Assoc. Lab. Anim. Sci. 2011, 50, 471–478. [Google Scholar]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb-prot5439. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Govero, J.; Smith, A.M.; Platt, D.J.; Fernandez, E.; Miner, J.J.; Diamond, M.S. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe 2016, 19, 720–730. [Google Scholar] [CrossRef] [Green Version]
- Gorman, M.J.; Caine, E.A.; Zaitsev, K.; Begley, M.C.; Weger-Lucarelli, J.; Uccellini, M.B.; Tripathi, S.; Morrison, J.; Yount, B.L.; Dinnon, K.H., 3rd; et al. An Immunocompetent Mouse Model of Zika Virus Infection. Cell Host Microbe 2018, 23, 672–685.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef] [Green Version]
- Manokaran, G.; Finol, E.; Wang, C.; Gunaratne, J.; Bahl, J.; Ong, E.Z.; Tan, H.C.; Sessions, O.M.; Ward, A.M.; Gubler, D.J.; et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 2015, 350, 217–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donald, C.L.; Brennan, B.; Cumberworth, S.L.; Rezelj, V.V.; Clark, J.J.; Cordeiro, M.T.; Freitas de Oliveira Franca, R.; Pena, L.J.; Wilkie, G.S.; Da Silva Filipe, A.; et al. Full Genome Sequence and sfRNA Interferon Antagonist Activity of Zika Virus from Recife, Brazil. PLoS Negl. Trop. Dis. 2016, 10, e0005048. [Google Scholar] [CrossRef] [PubMed]
- Bidet, K.; Dadlani, D.; Garcia-Blanco, M.A. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014, 10, e1004242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, V.K.; Griffin, D.E. Interferon gamma modulation of disease manifestation and the local antibody response to alphavirus encephalomyelitis. J. Gen. Virol. 2016, 97, 2908–2925. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Bramley, J.C.; Morosky, S.; Marques, E.T., Jr.; Cherry, S.; Sadovsky, Y.; Coyne, C.B. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe 2016, 19, 705–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, G.K.; Griffin, D.E. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science 2001, 293, 303–306. [Google Scholar] [CrossRef]
- Jagger, B.W.; Miner, J.J.; Cao, B.; Arora, N.; Smith, A.M.; Kovacs, A.; Mysorekar, I.U.; Coyne, C.B.; Diamond, M.S. Gestational Stage and IFN-lambda Signaling Regulate ZIKV Infection In Utero. Cell Host Microbe 2017, 22, 366–376.e3. [Google Scholar] [CrossRef] [Green Version]
- Baxter, V.K.; Griffin, D.E. Interferon-Gamma Modulation of the Local T Cell Response to Alphavirus Encephalomyelitis. Viruses 2020, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Caine, E.A.; Jagger, B.W.; Diamond, M.S. Animal Models of Zika Virus Infection during Pregnancy. Viruses 2018, 10, 598. [Google Scholar] [CrossRef] [Green Version]
- Carbaugh, D.L.; Zhou, S.; Sanders, W.; Moorman, N.J.; Swanstrom, R.; Lazear, H.M. Two genetic differences between closely-related Zika virus strains determine pathogenic outcome in mice. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Shan, C.; Muruato, A.E.; Jagger, B.W.; Richner, J.; Nunes, B.T.D.; Medeiros, D.B.A.; Xie, X.; Nunes, J.G.C.; Morabito, K.M.; Kong, W.P.; et al. A single-dose live-attenuated vaccine prevents Zika virus pregnancy transmission and testis damage. Nat. Commun. 2017, 8, 676. [Google Scholar] [CrossRef]
- Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Luo, H.; Xie, X.; Medeiros, D.B.A.; Wakamiya, M.; Tesh, R.B.; Barrett, A.D.; Wang, T.; et al. A live-attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat. Med. 2017, 23, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Xie, X.; Luo, H.; Shan, C.; Muruato, A.E.; Weaver, S.C.; Wang, T.; Shi, P.Y. A single-dose plasmid-launched live-attenuated Zika vaccine induces protective immunity. EBioMedicine 2018, 36, 92–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, A.; Tesh, R.B.; Wood, T.G.; Widen, S.G.; Ryman, K.D.; Barrett, A.D. Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing. J. Infect. Dis. 2014, 209, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Blaney, J.E., Jr.; Sathe, N.S.; Goddard, L.; Hanson, C.T.; Romero, T.A.; Hanley, K.A.; Murphy, B.R.; Whitehead, S.S. Dengue virus type 3 vaccine candidates generated by introduction of deletions in the 3’ untranslated region (3’-UTR) or by exchange of the DENV-3 3’-UTR with that of DENV-4. Vaccine 2008, 26, 817–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieft, J.S.; Rabe, J.L.; Chapman, E.G. New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation. RNA Biol. 2015, 12, 1169–1177. [Google Scholar] [CrossRef] [Green Version]
- Funk, A.; Truong, K.; Nagasaki, T.; Torres, S.; Floden, N.; Balmori Melian, E.; Edmonds, J.; Dong, H.; Shi, P.Y.; Khromykh, A.A. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 2010, 84, 11407–11417. [Google Scholar] [CrossRef] [Green Version]
- Villordo, S.M.; Filomatori, C.V.; Sanchez-Vargas, I.; Blair, C.D.; Gamarnik, A.V. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog. 2015, 11, e1004604. [Google Scholar] [CrossRef]
- Filomatori, C.V.; Carballeda, J.M.; Villordo, S.M.; Aguirre, S.; Pallares, H.M.; Maestre, A.M.; Sanchez-Vargas, I.; Blair, C.D.; Fabri, C.; Morales, M.A.; et al. Dengue virus genomic variation associated with mosquito adaptation defines the pattern of viral non-coding RNAs and fitness in human cells. PLoS Pathog. 2017, 13, e1006265. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sparks, H.; Monogue, B.; Akiyama, B.; Kieft, J.; Beckham, J.D. Disruption of Zika Virus xrRNA1-Dependent sfRNA1 Production Results in Tissue-Specific Attenuated Viral Replication. Viruses 2020, 12, 1177. https://doi.org/10.3390/v12101177
Sparks H, Monogue B, Akiyama B, Kieft J, Beckham JD. Disruption of Zika Virus xrRNA1-Dependent sfRNA1 Production Results in Tissue-Specific Attenuated Viral Replication. Viruses. 2020; 12(10):1177. https://doi.org/10.3390/v12101177
Chicago/Turabian StyleSparks, Hadrian, Brendan Monogue, Benjamin Akiyama, Jeffrey Kieft, and J. David Beckham. 2020. "Disruption of Zika Virus xrRNA1-Dependent sfRNA1 Production Results in Tissue-Specific Attenuated Viral Replication" Viruses 12, no. 10: 1177. https://doi.org/10.3390/v12101177
APA StyleSparks, H., Monogue, B., Akiyama, B., Kieft, J., & Beckham, J. D. (2020). Disruption of Zika Virus xrRNA1-Dependent sfRNA1 Production Results in Tissue-Specific Attenuated Viral Replication. Viruses, 12(10), 1177. https://doi.org/10.3390/v12101177





