Abstract
The ability to detect and respond to varying oxygen tension is an essential prerequisite to life. Several mechanisms regulate the cellular response to oxygen including the prolyl hydroxylase domain (PHD)/factor inhibiting HIF (FIH)-hypoxia inducible factor (HIF) pathway, cysteamine (2-aminoethanethiol) dioxygenase (ADO) system, and the lysine-specific demethylases (KDM) 5A and KDM6A. Using a systems-based approach we discuss the literature on oxygen sensing pathways in the context of virus replication in different tissues that experience variable oxygen tension. Current information supports a model where the PHD-HIF pathway enhances the replication of viruses infecting tissues under low oxygen, however, the reverse is true for viruses with a selective tropism for higher oxygen environments. Differences in oxygen tension and associated HIF signaling may play an important role in viral tropism and pathogenesis. Thus, pharmaceutical agents that modulate HIF activity could provide novel treatment options for viral infections and associated pathological conditions.
1. Introduction
Oxygen is essential for survival and organisms have developed mechanisms to detect and respond to variable oxygen tensions. These include the prolyl hydroxylase domain (PHD)/factor inhibiting HIF (FIH)-hypoxia inducible factor (HIF) pathway [1,2] along with the more recently described cysteamine (2-aminoethanethiol) dioxygenase (ADO) pathway [3] and lysine-specific demethylase (KDM) 5A and KDM6A pathways [4,5] (Figure 1). PHDs and KDMs are members of a family of enzymes that are dependent on oxygen, Fe(II), ascorbate and the Krebs cycle intermediate 2-oxoglutarate (2OG), that regulate fundamental cellular processes by catalysing the hydroxylation or demethylation of DNA, RNA or proteins such as histones (reviewed in [6]).
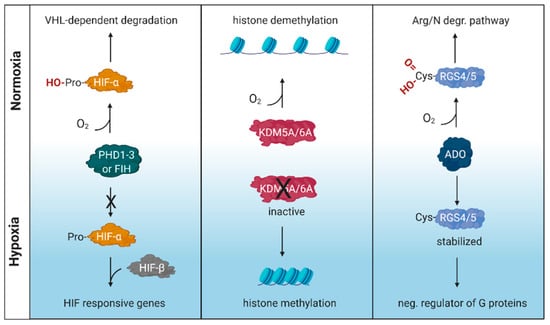
Figure 1.
Schematic illustration of oxygen sensing mechanisms. These include the prolyl hydroxylase domain (PHD)/factor inhibiting HIF (FIH)-hypoxia inducible factor (HIF) pathway, lysine-specific demethylase (KDM) 5A and KDM6A pathways and the cysteamine (2-aminoethanethiol) dioxygenase (ADO) pathway. RGS, regulator of G protein signaling; VHL, von Hippel–Lindau tumor suppressor gene. Created with BioRender.com.
The importance of HIFs in oxygen sensing was recognised by the 2019 Nobel Prize in Physiology or Medicine awarded to William Kaelin, Peter Ratcliffe and Gregg L. Semenza [7]. HIFs are heterodimeric transcription factors comprising an alpha and beta subunit that bind a consensus RCGTG(C) motif or hypoxic responsive element (HRE) in the promoter and enhancer regions of target genes. When oxygen is abundant, newly synthesised HIFα subunits, including HIF-1α and HIF-2α isomers, are hydroxylated by PHD or FIH, poly-ubiquitinated via von Hippel-Lindau factor (VHL) and targeted for proteasomal degradation. However, when oxygen is limiting these subunits are stabilised, translocate to the nucleus and dimerise with HIF-β to regulate a myriad of host target genes [1]. The PHD-HIF pathway is known to regulate genes with wide ranging functions, from metabolism and immunity, to DNA repair and carcinogenesis [8].
As obligatory intracellular parasites, viruses depend on the host cellular machinery to replicate. Important factors that shape the cellular microenvironment include temperature, pH and oxygen tension. As variable oxygen tension regulates host gene expression, protein modification, metabolism and epigenetic regulation, it is not surprising that oxygen availability can influence multiple steps in the viral life cycle [4,9]. A wide range of oxygen tensions are found within different tissues and organs, ranging from <1% in the skin to 14.5% in arterial blood vessels (reviewed in [10]). However, the majority of in vitro studies on viral replication have been conducted at comparatively high atmospheric oxygen tension (18%) [11], where HIFs are not active and consequently their role in viral replication may have been overlooked. Viral infection, inflammatory responses and tissue damage induced reactive oxygen species (ROS) can all stabilise HIF expression, highlighting the complex interplay between oxygen sensing pathways and viral replication [12,13]. Viruses have either DNA or RNA genomes [14] and this can define whether HIFs regulate viral transcription by direct binding to the viral genome or via indirect regulation of host genes. Screening DNA genomes representing the major viral families shows an approximate 50-fold variation in the frequency of HREs relative to the number of open reading frames (ORFs), ranging from 0.3 for HIV-1 (3 HRE/10 ORFs) to 14.7 for Molluscum contagiosum (2395 HRE/163 ORFs) (Figure 2). These data are consistent with variable sensitivity of viruses to low oxygen environments.
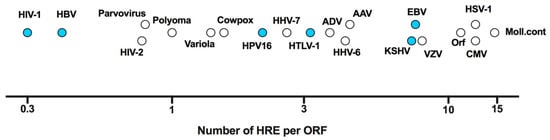
Figure 2.
Frequency of HIF response elements (HRE) in viral DNA genomes. The frequency of HRE elements (RCGTG) is plotted against the number of open reading frames (ORF) for a range of DNA viruses with those marked in blue discussed in this review. Referent sequences presented were selected from Genbank as follows (accession numbers in brackets); adeno-associated virus (AAV, NC_001401.2), adenovirus (ADV, AC_000007.1), cytomegalovirus (CMV, KU317610.1), cowpox virus (CowPox, NC_003663.2), Epstein–Barr virus (EBV, NC_009334.1), hepatitis B virus (HBV, NC_003977.2), human herpesvirus 6 (HHV-6, NC_000898.1), HHV-7 (NC_001716.2), Kaposi’s sarcoma-associated herpesvirus (KSHV, NC_009333.1), human immunodeficiency virus type 1 (HIV-1, NC_001802.1), HIV-2 (NC_001722.1), human papillomavirus 16 (HPV16, NC_001526.4), herpes simplex virus type 1 (HSV-1, NC_001806.2), human T-lymphotropic virus 1 (HTLV-1, NC_001436.1), Molluscum Contagiosum (Moll.cont, NC_001731.1), Orf virus (Orf, NC_005336.1), parvovirus (NC_000883.2), polyomavirus (NC_031757.1), varicella-zoster virus (VZV, NC_001348.1) and variola virus (NC_001611.1).
These data prompted us to review the current knowledge on how oxygen tension impacts viral replication (Figure 3) and how viruses manipulate the PHD-HIF pathway. Using a systems-based approach we discuss the literature in the context of viruses infecting and replicating in different tissue sites. Previous reviews have generally focused on the role of HIFs in viral carcinogenesis and host immunity [15,16,17,18]. Finally, we review pharmaceutical agents that modulate HIF activity and discuss their potential use as novel treatments for viral infections and associated pathological conditions [19,20,21].
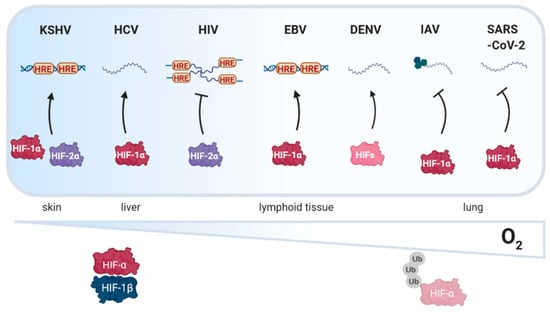
Figure 3.
Schematic illustration depicting how oxygen tension in different tissues affects viral replication. Hypoxia inducible factor (HIF) signaling enhances (black arrow) the replication of Kaposi’s sarcoma associated herpesvirus (KSHV), hepatitis C virus (HCV), Epstein Barr virus (EBV) and dengue virus (DENV). In contrast, human immunodeficiency virus type I (HIV-1), influenza A virus (IAV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication is dampened by HIF signaling. HRE, HIF response element, Ub, ubiquitin. Created with BioRender.com.
2. The Skin and Epidermis
The skin plays an essential role in responding to environmental stimuli, where specific deletion of HIF-1α from the epidermis in mice inhibits renal erythropoietin (EPO) synthesis in response to hypoxia [22]. Furthermore, mice with an epidermal deletion of the VHL factor, a negative regulator of HIF, show increased EPO expression and polycythemia.
2.1. Kaposi’s Sarcoma Associated Herpesvirus (KSHV)
KSHV is a member of the Herpesviridae double-stranded DNA viruses that infects epithelial cells and keratinocytes, where the local oxygen tension is in the range of 1–2.5% [10,23,24]. KSHV causes Kaposi’s sarcoma, the most common acquired immunodeficiency syndrome related neoplasm [25] and viral lytic gene activation is associated with oncogenesis [26]. In 2003, Haque et al. identified functional HREs in the promoter regions of the ORF34 and Rta genes, which are involved in lytic switch, late stage gene expression and viral particle production [27,28]. Hypoxia activated both promoters and while ORF34 promoter was regulated by both HIF-1α and HIF-2α, the Rta promoter was preferentially regulated by HIF-2α. In 2006, Haque et al. went on to identify an additional HRE in the ORF35–37 promoter region that was co-regulated by both HIF-1α and HIF-2α [29]. These active HREs in the KSHV lytic gene promoters support an essential role for HIFs in regulating viral latency. KSHV-associated tumours are highly vascularised, a process which is partly mediated by virus-induced HIF signaling. Firstly, KSHV encoded G protein-coupled receptor (vGPCR) induces vascular endothelial growth factor (VEGF) expression and associated angiogenesis [25]. The vGPCR induces HIF-1α regulatory domain phosphorylation via p38/MAPK and mTOR pathways [30]. Secondly, the KSHV-encoded viral interferon (IFN) regulatory factor 3 (vIFR3) binds to HIF-1α and prevents its degradation [31]. Finally, the KSHV encoded latency-associated nuclear antigen acts as a transcriptional coactivator interacting with HIF-1α protein and enhances HIF-1α mRNA transcription [32,33]. These studies are consistent with the reported 34% overlap between KSHV infection and hypoxia gene expression profiles [34], suggesting that KSHV exploits the HIF signaling pathway and multiple redundant pathways to ensure HIF expression. Furthermore, inhibiting HIF in immunodeficient athymic mice reduced VEGF expression and tumour growth in a syngeneic mouse model [30]. KSHV-associated angiogenesis was potentiated by the hypoxia mimetic, deferoxamine mesylate [35], demonstrating an essential role for HIFs in KSHV pathogenesis. KSHV can also cause non-skin related cancers, however, the role of HIFs in these non-skin related cancers have not been studied.
2.2. Human Papilloma Virus (HPV)
HPV are a family of small double-stranded DNA viruses that infect basal epithelia and specific strains have been associated with a risk of oncogenesis [36,37]. The link between HPV strains 16 and 18 and cervical cancer has led to their classification as high risk and integration of HPV16 associates with HIF-1α overexpression [38]. HPV16 and 18 encoded E6 and E7 proteins stabilise HIF-1α and induce VEGF and IL-8 expression that is associated with increased angiogenesis [39,40,41]. Three studies have suggested alternative non-exclusive mechanisms for HPV to stabilise HIFs: (1) E6 prevents HIF-1α association with VHL [42]; (2) E6 and E7 induce HIF-2α protein via liver kinase B1 (LKB1) modulation [43] and (3) E2 binding to mitochondrial membrane components of the respiratory chain induce reactive oxygen species (ROS) [44]. There are limited clinical studies addressing the translational impact of HIFs in HPV associated cancers. However, in a cohort of patients with oropharyngeal squamous cell carcinoma there was some evidence for HIF-1α expression in the tumour associating with clinical outcome [45]. Jo et al. reported that elevated VEGF levels in a cohort of patients diagnosed with HPV16-associated oropharynx squamous cell carcinoma was independent of HIF-1α expression [46]. Given the technical limitations in staining for HIF-1α in tissue [47] and the oxygen rich environment of the oropharynx, ex vivo patient sample processing methods may require optimisation before interpreting these studies. Further clinical studies are required to fully examine the relationship between HPV, HIFs and patient outcomes.
3. The Liver
The liver is a naturally low oxygen environment with the highest oxygen tension near its periportal region at 8%, gradually decreasing to 4% in the pericentral area. A recent single cell sequencing study of the mouse liver [48] reported this oxygen gradient to associate with liver zonation, a phenomenon where hepatocytes show distinct functional and structural heterogeneity across the liver [49]. These infections are the leading cause for HCC worldwide and associated with significant mortality, accounting for more than 1.3 million deaths per year [50]. Hepatitis B and C viruses are a global health problem causing acute and chronic infections that can lead to liver cirrhosis and hepatocellular carcinoma (HCC).
3.1. Hepatitis C Virus (HCV)
HCV is a positive-sense single-stranded RNA virus in the Flaviviridae family that infects the liver. Low oxygen has been reported to increase HCV RNA levels in human hepatoma Huh-7 and HepG2 model systems [51,52]. These studies reported differences in the HIF-dependency of the low oxygen-increase in HCV RNA that most likely reflects the different oxygen tensions used in the experimental models: Vassilaki et al. [51] cultured the infected cells at 3% oxygen and showed a minimal role for HIFs in regulating HCV replication, whereas Wilson et al. [52] reported a positive role for HIFs in regulating HCV replication at 1% oxygen. Since the HIF-PHD and FIH degradation pathways are still functionally active at 3% oxygen there will be limited HIF transcriptional activity, highlighting the importance of the % oxygen applied in vitro model systems and how this impacts on HIF signalling. The study by Vassilaki highlights the role for hydroxylases within the 2OG oxygenase family in regulating HCV replication. A recent report showed the importance of N6-methyladenosine (m6A) modification in the replication of HCV and other Flaviviridae RNAs and viral particle genesis that was mediated by fat mass and obesity-associated protein (FTO), a member of the 2OG oxygenases [53].
The phospholipase autotaxin is hypoxic regulated and pharmacological inhibition or siRNA silencing of autotaxin-lysophosphatidic acid signaling reduced HCV replication, suggesting a role for HIFs to promote HCV replication via this pathway [54]. Autotaxin generates the biologically active lipid lysophosphatidic acid that plays a pro-tumorigenic role in a wide number of cancers. Farquhar et al. reported a positive association between hypoxic gene expression in HCV associated HCC tissue and autotaxin transcript levels [54]. Furthermore, the HCV encoded core [55,56,57,58] and E1E2 glycoproteins [52] stabilised HIF expression by inducing an unfolded protein-stress response. Further reports showing that HCV induction of NF-kB, STAT-3, PI3-K-aKT, and p42/44 MAPK signaling stabilised HIF-1α [54,59,60,61], highlight the multiple pathways for HCV induced HIF signaling. Nassimuzzaman et al. [61] reported HIF-dependent VEGF expression in HCV infected cells and suggested a role for HCV in activating angiogenesis. This observation was extended by Wilson et al. who showed that HCV-induced VEGF reduced hepatoma cell polarity and potentiated viral transmission [62] and increased epithelial-mesenchymal transition, suggesting a role for HCV stabilised HIFs in promoting fibrosis and liver injury.
3.2. Hepatitis B Virus (HBV)
HBV is a member of the Hepadnaviridae family of partially double-stranded DNA viruses that infect the liver. In contrast to our knowledge of HIFs in regulating HCV, currently there are no published studies exploring the role of HIFs in regulating HBV transcription. Hallez et al. reported that hypoxia induced human deoxyribonuclease 1 (DNASE1) could catabolise the encapsidated DNA genomes, resulting in a high frequency of empty or ‘light’ virions HBV [63]. The majority of experiments used cobalt chloride and dimethyloxalylglycine, mimetic agents that can inhibit prolyl hydroxylase enzymes and stabilise HIFs. However, these mimetics have been reported to induce additional oxygen-independent biological effects [64,65].
Several reports have studied the HBV encoded regulatory protein HBx that interacts with a myriad of host factors including HIF-1α and contributes to liver pathogenesis. Yoo et al. reported that HBx induced HIF-1α protein and mRNA upregulation through two independent mechanisms: (1) upregulation of MTA1 and HDAC1/2 that perturbed HIF-1α deacetylation and (2) prevention of HIF-1α association with VHL [66,67,68]. HBx was also reported to bind HIF-1α and increase its stability [69] and C-terminal truncation of HBx abrogated this association [70]. A significant limitation of these studies is their dependence on HBx overexpression systems. Lui et al. reported that a variety of in vitro and in vivo HBV model systems failed to show a role for HBx in stabilizing or modifying HIF transcriptional activity However, increased HIF target gene expression was observed in liver tissue from a chronic hepatitis B (CHB) cohort that associated with inflammatory immune responses [71]. Since inflammation and associated oxidative stress are known to induce HIF-transcription, the authors conclude that HBV-associated inflammation drives HIF expression.
Several clinical studies have reported an association between increased HIF expression and HBV-related disease outcome. Xie et al. showed that HIF-1α expression in HCC associated with shorter survival [72]. Similarly, Osman et al. demonstrated that HIF-1α expression in HCC associated with larger tumour size, multifocal malignancies and more advanced disease [73]. Genetic variation in HIF-1α has also been linked with HBV-HCC risk, where the CG haplotype associated with increased risk relative to the CA haplotype [74]. Furthermore, a HIF-2α single nucleotide polymorphism rs13419896 associated with an increased risk of liver cirrhosis [75]. Similarly, high KDM5B expression showed a negative association with HCC prognosis [76]. These clinical studies support translational studies to explore the application of HIF inhibitors to HBV related pathologies.
4. The Immune System
Lymphoid organs operate at oxygen levels in the range of 0.5–4.5% O2 [77,78,79], which are sufficient to activate the HIF pathway [80].
4.1. Human Immunodeficiency Virus Type I (HIV-1)
Human immunodeficiency virus type I (HIV-1) is a retrovirus with an RNA genome that is reverse transcribed into cDNA and integrated into the host genome where it serves as a transcriptional template. If the integrated provirus is methylated and transcriptionally silenced, the viral genome becomes latent. Reactivation of HIV from latent reservoirs is a crucial step of ‘shock-and-kill’ treatment approaches. HIV-1 primarily replicates in CD4+ T cells and the major virus reservoirs are thought to be in lymphoid tissues: hence this virus has evolved to replicate in cells that may experience widely differing oxygen tensions. Accumulating evidence shows that a hypoxic environment inhibits HIV replication and reactivation [81,82]. HIF-2α can repress HIV transcription under low oxygen conditions (1% O2) via a direct interaction with a conserved HRE in the U3 region of the long terminal repeat (LTR) [81]. In addition, hypoxic reduction in cyclin T1 activity was reported to reduce Tat mediated transcription [82]. Duette et al. reported the hypoxia mimetic cobalt chloride induced a modest 1.5 fold increase in HIV-1 replication [83], however, since these mimetics activate additional signaling pathways [64,65] further investigations are required to explore these differences.
Chronic immune activation, metabolic changes and inflammation during HIV-1 replication induce mitochondrial ROS [83]. The HIV-1 accessory protein Vpr was reported to increase HIF-1α expression via induction of ROS [84] and is consistent with an independent report showing that expression of Vpr in the monocytic cell line U937 increased HIF-dependent glycolysis [85]. In line with these findings, clinical studies reported HIF-1α expression in the brain of AIDS patients diagnosed with dementia [84] and increased HIF-2α and VEGF protein in kidney biopsies from patients with HIV-associated nephropathy [86]. Taken together, these data suggest that the hypoxic environment may be relevant to any successful shock-and-kill antiviral strategy.
4.2. Human T-Lymphotropic Virus Type 1 (HTLV-1)
The human T-lymphotropic virus type 1 (HTLV-1) is a retrovirus that integrates into the host genome. An estimated 5–10 million people worldwide are infected by HTLV-1 and in 5–10% of cases this leads to a CD4-T cell malignancy, known as adult T-cell leukemia or a progressive inflammatory disease of the spinal cord. In contrast to HIV-1, hypoxia enhanced HTLV-1 reactivation from latency via a HIF-independent process that involved the 2OG metabolite, suggesting a role for oxygenases in reactivating the genome [87]. Furthermore, there is evidence that the HLTV-1 encoded transactivator protein, Tax, induces HIF-1α protein expression and suppresses expression of the cellular proapoptotic BH3-only proteins Bim and Bid [88]. Since HTLV-1 may benefit from a hypoxic environment through several different mechanisms, a better understanding of these interactions could identify therapeutic targets.
4.3. Epstein Barr Virus (EBV)
Epstein Barr virus (EBV) is a double-stranded DNA virus in the family of Herpesviridae that replicates in CD20+ B cells and epithelial cells of the oropharynx and is associated with a number of malignancies, including Hodgkin’s lymphoma and Burkett’s Lymphoma [89]. As described for KSHV, HIF-1α directly binds the primary latent-lytic switch BZLF1 gene promoter, Zp, and activates transcription [90,91]. The EBV latent membrane protein 1 (LMP1), an oncogenic protein capable of B-cell immortalisation, increases HIF-1α protein expression by upregulating the Siah1 E3 ubiquitin ligase that degrades PHD1 and PHD3 [89,92,93,94]. In addition, the loss of VHL/HIF-1α complexes stabilises HIF-1α and associated HIF signaling, including pyruvate dehydrogenase kinase 1 (PDK1) and the pyruvate kinase M2 (PKM2) isoform, resulting in an increase in lactate production and glucose consumption [95].
There is limited evidence of an association between the survival of patients with EBV+ nasopharyngeal cancers and HIF-1α or VEGF expression [96]. Although LMP1 expression was noted in all EBV+ nasopharyngeal carcinomas, there was limited evidence of LMP1 co-localisation with HIF-1α. Despite the lack of a direct correlation between HIF expression and nasopharyngeal cancer clinical outcome, Yang et al. reported that LMP1 blockade increased the sensitivity of nasopharyngeal carcinoma to radiotherapy by downregulating HIF-1α and VEGF activity and decreasing phosphorylated JNKs/c-Jun signaling [97]. This observation was further validated through an in vivo experiment showing a significant reduction in tumour growth.
4.4. Dengue Virus (DENV)
DENV is a member of the Flavivirus genus that includes yellow fever, Zika, West Nile and Japanese encephalitis viruses. DENV is the most prevalent arbovirus disease in the world, with a spectrum of symptoms ranging from asymptomatic to severe haemorrhagic fever. DENV is transmitted by Aedes aegypti mosquito with potentially 3.9 billion people currently at risk of contracting infection [98]. Severe dengue fever most often occurs in secondary/tertiary infections and is thought to be mediated via antibody-dependent enhanced (ADE) infection along with a skewed T cell response. DENV primarily replicates in macrophages [99,100] and infection is associated with an oxidative stress response that plays a role in the immunopathology of the disease [101]. Gan et al. showed that hypoxia enhanced antibody-dependent DENV infection of THP-1 cells and primary human monocytes by two independent mechanisms [102]. Firstly, HIF-1α upregulated fragment crystallisable gamma receptor IIA (FcγRIIA). Secondly, membranous lipid concentrations were increased under hypoxia independently of HIF-1α. These processes increased antibody-opsonised DENV infection of monocytes. HIFs were also reported to promote DENV RNA replication and translation through a HIF-1α dependent mechanism in Huh-7 hepatoma cells [103]. DENV induces an oxidative stress response that may lead to HIF expression and hypoxic reprogramming [104]. Frakolaki et al. reported that DENV infection activated a HRE-luciferase reporter plasmid in Huh7 cells under atmospheric oxygen tension [103]. However, no further experiments were conducted to directly examine HIF protein levels or transcription of HIF target genes, limiting the conclusions of the study. Further studies are required to dissect the influence of physiological oxygen tension and tissue specific mechanisms on DENV regulation as well as the influence of DENV replication on local oxygen tension and metabolism.
5. The Respiratory Tract
Oxygen levels in the lung can vary in different locations within adults but also during development. Le et al. reported a median oxygen tension of 5.6% for adult lung tissue [105]. HIFs play an important role in fetal lung development and when the lung is hypoxic, loss of HIF expression can lead to impaired lung development or even death [106]. At the same time HIF upregulation due to lung damage, pulmonary hypertension or in lung cancer can negatively correlate with disease outcome.
5.1. Influenza Virus (IAV)
Influenza A virus (IAV) is a negative-sense single-stranded RNA virus associated with respiratory infections. IAV strains differ in their preference to infect the upper or lower respiratory tract [107]. Zhao et al. reported that IAV H1N1 (PR8) replicated to a higher titre in mice with targeted HIF-1α knockout in type II alveolar epithelial cells (AEC2) [108], accompanied by increased lung inflammation and mortality. Although the underlying mechanisms require further exploration, these data support an indirect role for HIF-1α in regulating IAV replication by inhibiting autophagy as a result of decelerated glycolysis. IAV has been reported to increase glycolysis by enhancing glucose uptake, lactate production and oxygen consumption rates [109,110]. Oral treatment with BEZ235 (a putative PI3 K/mTOR inhibitor) decreased glycolysis and reduced virus replication and mortality in a mouse model [111]. This observation contrasts with data from Zhao et al. [108] that may reflect cell-type dependent metabolic responses in the lung. It is likely that the role of HIF in influenza infection is more complicated as Zhang et al. recently showed that RIG-I like receptor (RLR) activation, a key sensor of IAV infection, suppresses glycolysis by inhibiting hexokinase [112]. HIF-1α is an important regulator of metabolic processes and is potentially regulated by immune responses against the virus. Indeed, HIF-1α induces proinflammatory cytokines and plays a key role in AEC2 differentiation and alveolar repair following IAV infection through activation of Notch signaling [113,114,115].
Supplemental oxygen (hyperoxia) given to premature infants associates with an increased risk of lung dysfunction and susceptibility to respiratory infections in adult life [116,117,118]. In a mouse model hyperoxia caused a significant reduction of pro-surfactant protein C-positive alveolar epithelial Type II cells by 8 weeks of age [116]. This finding is supported by a report identifying a protective role of superoxide dismutase to hyperoxia in a transgenic mouse model. Hence, oxygen tension and metabolic changes can impact the alveolar epithelial balance and response to injury. Further studies would elucidate the metabolic effects of HIF on different cell types in the lung, IAV replication, and whether oxygen tension affects IAV tissue tropism.
Infection with IAV leads to acute lung injury which activates hypoxia signaling and HIF induction. Several groups reported that IAV infection stabilises HIF-1α, although their proposed mechanisms vary. Zhao et al. showed an increase in HIF-1α expression in H1N1 (PR8) infected A549 cells with a modest increase in mRNA levels [108]. Ren et al. observed HIF-1α induction during H1N1 (PR8) infection as a result of reduced FIH expression [119]. Huo et al. reported no increase in HIF mRNA or protein levels, but an increase in nuclear translocation. Since IAV infection can result in a caspase-dependent enlargement of nuclear pores and non-specific protein uptake by the nucleus, this finding requires further validation [120]. Infection of the murine mastocytoma cell line P815 with different IAV strains (human H1N1, avian H5N1 and H7N2) showed that HIF-1α was activated by H7N2 but not by H1N1 or H5N1 [121], suggesting differing abilities of IAV strains to activate HIF-signalling pathways. It was interesting to note that H7N2 infection which activated HIF-signaling replicated to lower titre. Given the known effect of HIF-1α on degranulation and inflammatory factor production in immune cells, these differences could have wide-ranging implications [122]. HIF signaling is likely to be an important determinant for the outcome and recovery from IAV infections.
5.2. SARS-Coronavirus-2 (SARS-CoV-2)
SARS-CoV-2 is a positive-sense single-stranded RNA virus in the Coronaviridae family and the causal agent of coronavirus disease 2019 (COVID-19) [123]. As of September 2020, over 30 million people have been infected by SARS-CoV-2 with 1 million fatalities. SARS-CoV-2 primarily targets the respiratory tract [124] and can result in pneumonia and severe acute respiratory distress syndrome especially in the elderly and in individuals with comorbidities [124,125]. Epithelial cell death and lung inflammation are major hallmarks of SARS-CoV-2 induced tissue damage [126]. Although the clinical presentation is heterogeneous, with many mild cases, a defining feature of severe COVID-19 is a marked hypoxaemia. Recent studies show multi-organ involvement in severe COVID-19 disease [127], including the gastrointestinal tract [128] and central nervous system [127,128,129]. SARS-CoV-2 encoded Spike protein binds human angiotensin-converting enzyme (ACE2) and the transmembrane proteases [130,131], serine 2 (TMPRSS2) and furin, trigger the fusion of viral and cell membranes [130,131]. Hypoxia has been reported to reduce ACE2 expression in lung pulmonary arterial smooth muscle cells [132] and haematopoietic stem cell precursors [133] via regulating ACE1. Wing et al. showed that hypoxia and the HIF PHD inhibitor Roxadustat reduced ACE2 expression and inhibited SARS-CoV-2 entry in lung epithelial cells via a HIF-1α dependent signalling pathway [134]. Importantly, this study showed that hypoxia and pharmacological activation of HIFs inhibited SARS-CoV-2 RNA replication, showing that post-entry steps in the viral life cycle are oxygen-sensitive. This is consistent with a recent report showing a gradient of ACE2 expression in proximal (high) versus distal (low) pulmonary epithelial cells that associates with SARS-CoV-2 infection [124]. In contrast, neonatal hyperoxia increases ACE2 and TMPRSS2 expression in an age-dependent fashion [135] via a loss of type II alveolar epithelial cells. Given reports that neonatal hyperoxia associates with risk of more severe IAV infection in adults via a loss of type II alveolar epithelial cells [113,114,115], this provides an explanation for the increased severity of COVID-19 in elderly and people with pre-existing co-morbidities.
HIF-1α protein was detected in monocytes isolated from bronchoalveolar lavage and circulating neutrophils in COVID-19 patients [136,137]. These observations were validated in vitro and showed that SARS-CoV-2 infection of monocytes induced mitochondrial ROS and stabilised HIF-1α expression and associated gene transcription [136]. SARS-CoV-2 infection of human brain organoids also showed evidence of virus-dependent HIF expression [138]. In contrast, Appelberg et al. reported that SARS-CoV-2 infection of human hepatoma Huh-7 cells repressed HIF-1α expression [139], suggesting cell-type specific effects of viral infection on this pathway. Codo et al. demonstrated that pharmacological inhibition of mitochondrial ROS and associated HIF-1α activity in monocytes reduced SARS-CoV-2 infection and interleukin-1β expression in monocytes [136]. Furthermore, inhibition of HIF-1α restored T cell proliferation and rescued apoptosis in co-cultured A549 cells. These data contrast to those reported by Wing et al. who showed that treatment of lung epithelial cells with the HIF PHD inhibitor Roxadustat significantly reduced SARS-CoV-2 entry and replication [134]. These contrasting observations may reflect cell-type specific differences; for example, monocytes have limited permissivity to support SARS-CoV-2 replication and viral RNA levels were substantially lower than reported in lung epithelial cells. Collectively, these studies highlight the importance of hypoxia and HIF signalling in multiple aspects of SARS-CoV-2 life cycle, suggesting that targeting the HIF oxygen sensing pathway could offer a novel therapeutic modality for COVID-19. The diverse role of HIFs and other hallmarks of a low oxygen environment in the inflamed lung are likely to be complex and are worthy of further investigation.
6. Therapeutic Implications
6.1. HIF Modifiers
Given the important role that the PHD-HIF pathway plays in regulating the replication and associated pathogenesis of many viruses, inhibitors targeting PHD and HIF undergoing clinical trials could be repurposed for the treatment of viral diseases. Four PHD inhibitors that stabilise HIFα have been evaluated in phase II and III clinical trial studies for the treatment of anemia: Roxadustat (FG4592, FibroGen), Daprodustat (GSK1278863, GlaxoSmithKline), Molidustat (Bay8503934, Bayer), Vadadustat (AKB-6548, Akebia) [20]. Roxadustat was recently licensed for the treatment of chronic kidney disease and anemia in China [140]. Future studies could examine the therapeutic potential of these inhibitors in regulating viruses such as HIV and IAV that are repressed by HIFs.
A number of clinical trials have assessed the efficacy of HIF inhibitors in patients with advanced or refractory cancers [21]. The HIF-1α synthesis inhibitor Digoxin is undergoing phase II clinical trial for Kaposi’s Sarcoma [141]. CRLX101, a HIF-1α expression inhibitor, has completed a phase II trial in combination with bevacizumab, an antibody that targets VEGF, for the treatment of recurrent platinum-resistance ovarian, tubal, and primary peritoneal cancers and reduce tumor frequency and size [142,143]. However, a randomised phase II trial in patients with advanced renal carcinoma did not show any improvement in progression-free survival [144]. A phase II trial for CRLX101 in combination with capecitabine and radiotherapy for locally advanced rectal cancer is ongoing [145]. The HIF-2α specific dimerisation inhibitors PT2385 [146,147] and PT2977 [148,149] are being tested for the treatment of recurrent glioblastoma, non-metastatic VHL-associated and advanced clear cell renal carcinoma [149,150,151]. Viruses known to be positively regulated by HIF-2α, including KSHV and HCV could benefit from clinical trials focusing on HIF inhibitors. Furthermore, viruses whose pathogenesis has been linked to HIF-signalling, including HBV, HCV, HPV, EBV, KSHV and SARS-CoV-2 may benefit from HIF inhibitors. Since KDM5 demethylases induce a robust interferon response resulting in an increased resistant to infection, the first KDM5 inhibitor entered a phase I clinical trial for the treatment of HBV infection [152,153].
6.2. Engineered Oncolytic Viruses
In addition to direct targeting the PHD-HIF pathway, a number of studies have engineered oncolytic viruses to encode HRE to promote their replication in hypoxic tumour environments as anti-cancer agents. Post et al. inserted a HRE within the E1A promoter of the adenovirus genome and demonstrated enhanced viral-mediated cytolysis of human brain tumour cells under hypoxic conditions [154]. A similar study using an adenovirus expressing E1A under the control of a HRE showed preferential lysis of hepatoma, pancreatic cancer and lung tumour cell lines under hypoxic conditions [155]. The authors reported that the recombinant viruses resulted in a significant survival improvement in a nude mice xenograft model of prostate cancer. As hypoxia commonly occurs in solid tumours, such targeted genetic engineering of oncolytic viruses represents a promising novel cancer treatment modality.
Viruses that are used for vaccination purposes may also regulate HIF pathways. Vaccinia virus (VACV) has traditionally been used as a vaccine against smallpox and stabilise HIF expression via the C16 protein [156]. The N-terminal region of C16 binds and inhibits the human oxygen sensor, PHD2, resulting in HIF expression under normoxic conditions. Mazzon et al. compared the metabolic alterations of cells infected with wild type VACV or a mutant lacking C16 and found a role for C16 in regulating nucleotide, glucose and glutamine metabolic pathways [157]. C16 has homologs in other poxviruses, suggesting an evolutionary conserved role in their replication pathways [156]. Another example of a virus that modulates HIF signaling is the mammalian orthoreovirus [158,159,160,161]. This oncolytic virus has completed phase I-III clinical trials against many different types of cancer [162]; yet little is known about the specific mechanisms involved and further studies will greatly enrich therapeutic options.
7. Concluding Statement
This review has illustrated how variable oxygen tension in different tissues can affect many stages of the virus life cycle: regulating entry receptors, replication machinery, particle genesis and host-pathogen interactions (Table 1). Current information suggests that hypoxia preferentially enhances the replication of viruses with a tropism for low oxygen environments, whereas HIFs can dampen the replication of viruses that replicate in tissues with higher oxygen levels. Differences in oxygen tension and associated HIF signaling may play an important role in viral tropism and pathogenesis. Viruses have evolved to replicate in a tissue specific oxygen environment and developed ways to manipulate the metabolic micro-environment to their advantage. Given their wide-ranging effects on cellular metabolism low oxygen environments may influence the efficacy of both antiviral agents and immune based therapies and is worthy of further study. In vitro cell-based systems that utilise physiological oxygen tension may provide improved pre-clinical models for evaluating new anti-viral agents. Further studies to understand these mechanisms are crucial for generating novel or repurposing existing treatment strategies for viral infection and pathogenesis.

Table 1.
Mechanisms in which viruses interact with HIFs.
Author Contributions
P.J.L. conducted the literature review and co-wrote the article, P.B. provided sequence analysis, J.A.M. edited and co-wrote the article and M.S. conducted the literature review and co-wrote the article. P.J.L. DPhil studies are funded by the Hoffmann Foundation. All authors have read and agreed to the published version of the manuscript.
Funding
The McKeating laboratory is funded by Wellcome Trust IA 200838/Z/16/Z, MRC project grant MR/R022011/1 and by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Science (CIFMS), China (grant number: 2018-I2M-2-002).
Acknowledgments
The authors would like to thank James Harris and Andrea Magri for their helpful comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pugh, C.W.; Ratcliffe, P.J. New horizons in hypoxia signaling pathways. Exp. Cell Res. 2017, 356, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, P.J. Oxygen sensing and hypoxia signalling pathways in animals: The implications of physiology for cancer. J. Physiol. 2013, 591, 2027–2042. [Google Scholar] [CrossRef] [PubMed]
- Masson, N.; Keeley, T.P.; Giuntoli, B.; White, M.D.; Puerta, M.L.; Perata, P.; Hopkinson, R.J.; Flashman, E.; Licausi, F.; Ratcliffe, P.J. Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 2019, 365, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.A.; Laukka, T.; Myllykoski, M.; Ringel, A.E.; Booker, M.A.; Tolstorukov, M.Y.; Meng, Y.J.; Meier, S.R.; Jennings, R.B.; Creech, A.L.; et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 2019, 363, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Batie, M.; Frost, J.; Frost, M.; Wilson, J.W.; Schofield, P.; Rocha, S. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 2019, 363, 1222–1226. [Google Scholar] [CrossRef]
- Ploumakis, A.; Coleman, M.L. OH, the Places You’ll Go! Hydroxylation, Gene Expression, and Cancer. Mol. Cell 2015, 58, 729–741. [Google Scholar] [CrossRef]
- Oxygen Sensing: After the Nobel. Cell 2020, 180, 7–8. [CrossRef] [PubMed]
- Santos, S.A.D.; Andrade, D.R.J. HIF-1alpha and infectious diseases: A new frontier for the development of new therapies. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e92. [Google Scholar] [CrossRef]
- Choudhry, H.; Harris, A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018, 27, 281–298. [Google Scholar] [CrossRef]
- Carreau, A.; El Hafny-Rahbi, B.; Matejuk, A.; Grillon, C.; Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef]
- Morinet, F.; Parent, M.; Pillet, S.; Koken, M.; Lebbé, C.; Capron, C. Hypoxia inducible factor one alpha and human viral pathogens. Curr. Res. Transl. Med. 2017, 65, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Masson, N.; Singleton, R.S.; Sekirnik, R.; Trudgian, D.C.; Ambrose, L.J.; Miranda, M.X.; Tian, Y.M.; Kessler, B.M.; Schofield, C.J.; Ratcliffe, P.J. The FIH hydroxylase is a cellular peroxide sensor that modulates HIF transcriptional activity. EMBO Rep. 2012, 13, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Bonello, S.; Zähringer, C.; BelAiba, R.S.; Djordjevic, T.; Hess, J.; Michiels, C.; Kietzmann, T.; Görlach, A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Durmuş, S.; Ülgen, K. Comparative interactomics for virus-human protein-protein interactions: DNA viruses versus RNA viruses. FEBS Open Bio. 2017, 7, 96–107. [Google Scholar] [CrossRef]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef]
- Bhandari, T.; Nizet, V. Hypoxia-Inducible Factor (HIF) as a Pharmacological Target for Prevention and Treatment of Infectious Diseases. Infect Dis. Ther. 2014, 3, 159–174. [Google Scholar] [CrossRef]
- Cuninghame, S.; Jackson, R.; Zehbe, I. Hypoxia-inducible factor 1 and its role in viral carcinogenesis. Virology 2014, 456–457, 370–383. [Google Scholar] [CrossRef]
- Gan, E.S.; Ooi, E.E. Oxygen: Viral friend or foe? Virol. J. 2020, 17, 115. [Google Scholar] [CrossRef]
- Chan, M.C.; Holt-Martyn, J.P.; Schofield, C.J.; Ratcliffe, P.J. Pharmacological targeting of the HIF hydroxylases--A new field in medicine development. Mol. Aspects Med. 2016, 47–48, 54–75. [Google Scholar] [CrossRef]
- Singh, A.; Wilson, J.W.; Schofield, C.J.; Chen, R. Hypoxia-inducible factor (HIF) prolyl hydroxylase inhibitors induce autophagy and have a protective effect in an in-vitro ischaemia model. Sci. Rep. 2020, 10, 1597. [Google Scholar] [CrossRef]
- Fallah, J.; Rini, B.I. HIF Inhibitors: Status of Current Clinical Development. Curr. Oncol. Rep. 2019, 21, 6. [Google Scholar] [CrossRef]
- Boutin, A.T.; Weidemann, A.; Fu, Z.; Mesropian, L.; Gradin, K.; Jamora, C.; Wiesener, M.; Eckardt, K.U.; Koch, C.J.; Ellies, L.G.; et al. Epidermal sensing of oxygen is essential for systemic hypoxic response. Cell 2008, 133, 223–234. [Google Scholar] [CrossRef]
- Chakraborty, S.; Veettil, M.V.; Chandran, B. Kaposi’s Sarcoma Associated Herpesvirus Entry into Target Cells. Front. Microbiol. 2012, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Washington, A.T.; Singh, G.; Aiyar, A. Diametrically opposed effects of hypoxia and oxidative stress on two viral transactivators. Virol. J. 2010, 7, 93. [Google Scholar] [CrossRef]
- Sodhi, A.; Montaner, S.; Patel, V.; Zohar, M.; Bais, C.; Mesri, E.A.; Gutkind, J.S. The Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res. 2000, 60, 4873–4880. [Google Scholar]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Davis, D.A.; Wang, V.; Widmer, I.; Yarchoan, R. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) contains hypoxia response elements: Relevance to lytic induction by hypoxia. J. Virol. 2003, 77, 6761–6768. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Watanabe, T.; Yagi, S.; Yamanaka, T.; Fujimuro, M. Kaposi’s sarcoma-associated herpesvirus ORF34 is essential for late gene expression and virus production. Sci. Rep. 2017, 7, 329. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Wang, V.; Davis, D.A.; Zheng, Z.M.; Yarchoan, R. Genetic organization and hypoxic activation of the Kaposi’s sarcoma-associated herpesvirus ORF34-37 gene cluster. J. Virol. 2006, 80, 7037–7051. [Google Scholar] [CrossRef] [PubMed]
- Jham, B.C.; Ma, T.; Hu, J.; Chaisuparat, R.; Friedman, E.R.; Pandolfi, P.P.; Schneider, A.; Sodhi, A.; Montaner, S. Amplification of the angiogenic signal through the activation of the TSC/mTOR/HIF axis by the KSHV vGPCR in Kaposi’s sarcoma. PLoS ONE 2011, 6, e19103. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.C.; Joo, C.H.; Gack, M.U.; Lee, H.R.; Jung, J.U. Kaposi’s sarcoma-associated herpesvirus viral IFN regulatory factor 3 stabilizes hypoxia-inducible factor-1 alpha to induce vascular endothelial growth factor expression. Cancer Res. 2008, 68, 1751–1759. [Google Scholar] [CrossRef]
- Cai, Q.; Lan, K.; Verma, S.C.; Si, H.; Lin, D.; Robertson, E.S. Kaposi’s sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1 alpha to upregulate RTA expression during hypoxia: Latency control under low oxygen conditions. J. Virol. 2006, 80, 7965–7975. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Murakami, M.; Si, H.; Robertson, E.S. A potential alpha-helix motif in the amino terminus of LANA encoded by Kaposi’s sarcoma-associated herpesvirus is critical for nuclear accumulation of HIF-1alpha in normoxia. J. Virol. 2007, 81, 10413–10423. [Google Scholar] [CrossRef] [PubMed]
- Viollet, C.; Davis, D.A.; Tekeste, S.S.; Reczko, M.; Ziegelbauer, J.M.; Pezzella, F.; Ragoussis, J.; Yarchoan, R. RNA Sequencing Reveals that Kaposi Sarcoma-Associated Herpesvirus Infection Mimics Hypoxia Gene Expression Signature. PLoS Pathog. 2017, 13, e1006143. [Google Scholar] [CrossRef] [PubMed]
- Carroll, P.A.; Kenerson, H.L.; Yeung, R.S.; Lagunoff, M. Latent Kaposi’s sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J. Virol. 2006, 80, 10802–10812. [Google Scholar] [CrossRef]
- Schiffman, M.; Castle, P.E.; Jeronimo, J.; Rodriguez, A.C.; Wacholder, S. Human papillomavirus and cervical cancer. Lancet (London, England) 2007, 370, 890–907. [Google Scholar] [CrossRef]
- Bansal, A.; Singh, M.P.; Rai, B. Human papillomavirus-associated cancers: A growing global problem. Int. J. Appl. Basic Med. Res. 2016, 6, 84–89. [Google Scholar] [CrossRef]
- Kim, S.H.; Koo, B.S.; Kang, S.; Park, K.; Kim, H.; Lee, K.R.; Lee, M.J.; Kim, J.M.; Choi, E.C.; Cho, N.H. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int. J. Cancer 2007, 120, 1418–1425. [Google Scholar] [CrossRef]
- Li, G.; He, L.; Zhang, E.; Shi, J.; Zhang, Q.; Le, A.D.; Zhou, K.; Tang, X. Overexpression of human papillomavirus (HPV) type 16 oncoproteins promotes angiogenesis via enhancing HIF-1α and VEGF expression in non-small cell lung cancer cells. Cancer Lett. 2011, 311, 160–170. [Google Scholar] [CrossRef]
- He, L.; Zhang, E.; Shi, J.; Li, X.; Zhou, K.; Zhang, Q.; Le, A.D.; Tang, X. (−)-Epigallocatechin-3-gallate inhibits human papillomavirus (HPV)-16 oncoprotein-induced angiogenesis in non-small cell lung cancer cells by targeting HIF-1α. Cancer Chemother. Pharmcol. 2013, 71, 713–725. [Google Scholar] [CrossRef]
- Fan, R.; Hou, W.J.; Zhao, Y.J.; Liu, S.L.; Qiu, X.S.; Wang, E.H.; Wu, G.P. Overexpression of HPV16 E6/E7 mediated HIF-1α upregulation of GLUT1 expression in lung cancer cells. Tumour Biol. 2016, 37, 4655–4663. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Meng, X.; Ma, J.; Zheng, Y.; Wang, Q.; Wang, Y.; Shang, H. Human papillomavirus 16 E6 contributes HIF-1α induced Warburg effect by attenuating the VHL-HIF-1α interaction. Int. J. Mol. Sci. 2014, 15, 7974–7986. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.S.; Sun, J.; Wang, S.; Chung, K.; Du, J.T.; Wang, J.; Qiu, X.S.; Wang, E.H.; Wu, G.P. HPV16 E6/E7 upregulates HIF-2α and VEGF by inhibiting LKB1 in lung cancer cells. Tumour Biol. 2017, 39, 1010428317717137. [Google Scholar] [CrossRef]
- Lai, D.; Tan, C.L.; Gunaratne, J.; Quek, L.S.; Nei, W.; Thierry, F.; Bellanger, S. Localization of HPV-18 E2 at mitochondrial membranes induces ROS release and modulates host cell metabolism. PLoS ONE 2013, 8, e75625. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.; Zhang, M.; Veillard, A.S.; Jahanbani, J.; Lee, C.S.; Jones, D.; Harnett, G.; Clark, J.; Elliott, M.; Milross, C.; et al. The prognostic significance of hypoxia inducing factor 1-α in oropharyngeal cancer in relation to human papillomavirus status. Oral. Oncol. 2013, 49, 354–359. [Google Scholar] [CrossRef]
- Jo, S.; Juhasz, A.; Zhang, K.; Ruel, C.; Loera, S.; Wilczynski, S.P.; Yen, Y.; Liu, X.; Ellenhorn, J.; Lim, D.; et al. Human papillomavirus infection as a prognostic factor in oropharyngeal squamous cell carcinomas treated in a prospective phase II clinical trial. Anticancer Res. 2009, 29, 1467–1474. [Google Scholar] [CrossRef]
- Macklin, P.S.; Yamamoto, A.; Browning, L.; Hofer, M.; Adam, J.; Pugh, C.W. Recent advances in the biology of tumour hypoxia with relevance to diagnostic practice and tissue-based research. J. Pathol. 2020, 250, 593–611. [Google Scholar] [CrossRef]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Toth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017, 542, 352–356. [Google Scholar] [CrossRef]
- Jungermann, K.; Kietzmann, T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu. Rev. Nutr. 1996, 16, 179–203. [Google Scholar] [CrossRef]
- Ringelhan, M.; McKeating, J.A.; Protzer, U. Viral hepatitis and liver cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- Vassilaki, N.; Kalliampakou, K.I.; Kotta-Loizou, I.; Befani, C.; Liakos, P.; Simos, G.; Mentis, A.F.; Kalliaropoulos, A.; Doumba, P.P.; Smirlis, D.; et al. Low oxygen tension enhances hepatitis C virus replication. J. Virol. 2013, 87, 2935–2948. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.K.; Brimacombe, C.L.; Rowe, I.A.; Reynolds, G.M.; Fletcher, N.F.; Stamataki, Z.; Bhogal, R.H.; Simões, M.L.; Ashcroft, M.; Afford, S.C.; et al. A dual role for hypoxia inducible factor-1α in the hepatitis C virus lifecycle and hepatoma migration. J. Hepatol. 2012, 56, 803–809. [Google Scholar] [CrossRef]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, M.J.; Humphreys, I.S.; Rudge, S.A.; Wilson, G.K.; Bhattacharya, B.; Ciaccia, M.; Hu, K.; Zhang, Q.; Mailly, L.; Reynolds, G.M.; et al. Autotaxin-lysophosphatidic acid receptor signalling regulates hepatitis C virus replication. J. Hepatol. 2017, 66, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Selimovic, D.; Ghozlan, H.; Abdel-kader, O. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology 2009, 49, 1469–1482. [Google Scholar] [CrossRef]
- Abe, M.; Koga, H.; Yoshida, T.; Masuda, H.; Iwamoto, H.; Sakata, M.; Hanada, S.; Nakamura, T.; Taniguchi, E.; Kawaguchi, T.; et al. Hepatitis C virus core protein upregulates the expression of vascular endothelial growth factor via the nuclear factor-κB/hypoxia-inducible factor-1α axis under hypoxic conditions. Hepatol. Res. 2012, 42, 591–600. [Google Scholar] [CrossRef]
- Liu, X.H.; Zhou, X.; Zhu, C.L.; Song, H.; Liu, F. [Effects of HCV core protein on the expression of hypoxia-inducible factor 1 alpha and vascular endothelial growth factor]. Zhonghua Gan Zang Bing Za Zhi 2011, 19, 751–754. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, X.; Wang, S.; Yan, X.; Tang, Z.; Wu, K.; Li, Y.; Liu, F. Hepatitis C virus core protein induces hypoxia-inducible factor 1α-mediated vascular endothelial growth factor expression in Huh7.5.1 cells. Mol. Med. Rep. 2014, 9, 2010–2014. [Google Scholar] [CrossRef]
- Ripoli, M.; D’Aprile, A.; Quarato, G.; Sarasin-Filipowicz, M.; Gouttenoire, J.; Scrima, R.; Cela, O.; Boffoli, D.; Heim, M.H.; Moradpour, D.; et al. Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation. J. Virol. 2010, 84, 647–660. [Google Scholar] [CrossRef]
- Jung, G.S.; Jeon, J.H.; Choi, Y.K.; Jang, S.Y.; Park, S.Y.; Kim, S.W.; Byun, J.K.; Kim, M.K.; Lee, S.; Shin, E.C.; et al. Pyruvate dehydrogenase kinase regulates hepatitis C virus replication. Sci. Rep. 2016, 6, 30846. [Google Scholar] [CrossRef]
- Nasimuzzaman, M.; Waris, G.; Mikolon, D.; Stupack, D.G.; Siddiqui, A. Hepatitis C virus stabilizes hypoxia-inducible factor 1alpha and stimulates the synthesis of vascular endothelial growth factor. J. Virol. 2007, 81, 10249–10257. [Google Scholar] [CrossRef] [PubMed]
- Mee, C.J.; Farquhar, M.J.; Harris, H.J.; Hu, K.; Ramma, W.; Ahmed, A.; Maurel, P.; Bicknell, R.; Balfe, P.; McKeating, J.A. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology 2010, 138, 1134–1142. [Google Scholar] [CrossRef]
- Hallez, C.; Li, X.; Suspène, R.; Thiers, V.; Bouzidi, M.S.; Dorobantu, C.M.; Lucansky, V.; Wain-Hobson, S.; Gaudin, R.; Vartanian, J.-P. Hypoxia-induced human deoxyribonuclease I is a cellular restriction factor of hepatitis B virus. Nat. Microbiol. 2019, 4, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Triantafyllou, A.; Liakos, P.; Tsakalof, A.; Georgatsou, E.; Simos, G.; Bonanou, S. Cobalt induces hypoxia-inducible factor-1alpha (HIF-1alpha) in HeLa cells by an iron-independent, but ROS-, PI-3K- and MAPK-dependent mechanism. Free Radic. Res. 2006, 40, 847–856. [Google Scholar] [CrossRef]
- Zhigalova, N.; Artemov, A.; Mazur, A.; Prokhortchouk, E. Transcriptome sequencing revealed differences in the response of renal cancer cells to hypoxia and CoCl2 treatment. F1000Res 2015, 4, 1518. [Google Scholar] [CrossRef][Green Version]
- Yoo, Y.G.; Oh, S.H.; Park, E.S.; Cho, H.; Lee, N.; Park, H.; Kim, D.K.; Yu, D.Y.; Seong, J.K.; Lee, M.O. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J. Biol. Chem. 2003, 278, 39076–39084. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.G.; Cho, S.; Park, S.; Lee, M.O. The carboxy-terminus of the hepatitis B virus X protein is necessary and sufficient for the activation of hypoxia-inducible factor-1alpha. FEBS Lett. 2004, 577, 121–126. [Google Scholar] [CrossRef]
- Yoo, Y.G.; Na, T.Y.; Seo, H.W.; Seong, J.K.; Park, C.K.; Shin, Y.K.; Lee, M.O. Hepatitis B virus X protein induces the expression of MTA1 and HDAC1, which enhances hypoxia signaling in hepatocellular carcinoma cells. Oncogene 2008, 27, 3405–3413. [Google Scholar] [CrossRef] [PubMed]
- Moon, E.J.; Jeong, C.H.; Jeong, J.W.; Kim, K.R.; Yu, D.Y.; Murakami, S.; Kim, C.W.; Kim, K.W. Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia-inducible factor-1alpha. Faseb J. 2004, 18, 382–384. [Google Scholar] [CrossRef]
- Liu, L.P.; Hu, B.G.; Ye, C.; Ho, R.L.; Chen, G.G.; Lai, P.B. HBx mutants differentially affect the activation of hypoxia-inducible factor-1α in hepatocellular carcinoma. Br. J. Cancer 2014, 110, 1066–1073. [Google Scholar] [CrossRef]
- Liu, P.J.; Harris, J.M.; Marchi, E.; D’Arienzo, V.; Michler, T.; Wing, P.A.C.; Magri, A.; Ortega-Prieto, A.M.; van de Klundert, M.; Wettengel, J.; et al. Hypoxic gene expression in chronic hepatitis B virus infected patients is not observed in state-of-the-art in vitro and mouse infection models. Sci. Rep. 2020, 10, 14101. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Song, J.; Liu, K.; Ji, H.; Shen, H.; Hu, S.; Yang, G.; Du, Y.; Zou, X.; Jin, H.; et al. The expression of hypoxia-inducible factor-1alpha in hepatitis B virus-related hepatocellular carcinoma: Correlation with patients’ prognosis and hepatitis B virus X protein. Dig. Dis. Sci. 2008, 53, 3225–3233. [Google Scholar] [CrossRef] [PubMed]
- Osman, N.A.; Abd El-Rehim, D.M.; Kamal, I.M. Defective Beclin-1 and elevated hypoxia-inducible factor (HIF)-1α expression are closely linked to tumorigenesis, differentiation, and progression of hepatocellular carcinoma. Tumour Biol. 2015, 36, 4293–4299. [Google Scholar] [CrossRef]
- Liu, Y.; Sui, J.; Zhai, L.; Yang, S.; Huang, L.; Huang, L.; Mo, C.; Wu, J.; Li, S.; Qin, X. Genetic polymorphisms in hypoxia-inducible factor-1a gene and its association with HBV-related hepatocellular carcinoma in a Chinese population. Med. Oncol. 2014, 31, 200. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, C.; Deng, Y.; Liu, Y.; Zhao, J.; Huang, X.; Tang, W.; Sun, Y.; Qin, X.; Li, S. Association of Hypoxia-Inducible Factor-2 Alpha Gene Polymorphisms with the Risk of Hepatitis B Virus-Related Liver Disease in Guangxi Chinese: A Case-Control Study. PLoS ONE 2016, 11, e0158241. [Google Scholar] [CrossRef] [PubMed]
- Shigekawa, Y.; Hayami, S.; Ueno, M.; Miyamoto, A.; Suzaki, N.; Kawai, M.; Hirono, S.; Okada, K.I.; Hamamoto, R.; Yamaue, H. Overexpression of KDM5B/JARID1B is associated with poor prognosis in hepatocellular carcinoma. Oncotarget 2018, 9, 34320–34335. [Google Scholar] [CrossRef]
- Caldwell, C.C.; Kojima, H.; Lukashev, D.; Armstrong, J.; Farber, M.; Apasov, S.G.; Sitkovsky, M.V. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J. Immunol. 2001, 167, 6140–6149. [Google Scholar] [CrossRef] [PubMed]
- Braun, R.D.; Lanzen, J.L.; Snyder, S.A.; Dewhirst, M.W. Comparison of tumor and normal tissue oxygen tension measurements using OxyLite or microelectrodes in rodents. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H2533–H2544. [Google Scholar] [CrossRef]
- Hangai-Hoger, N.; Tsai, A.G.; Cabrales, P.; Intaglietta, M. Terminal lymphatics: The potential “lethal corner” in the distribution of tissue pO2. Lymphat. Res. Biol. 2007, 5, 159–168. [Google Scholar] [CrossRef]
- Ohta, A.; Diwanji, R.; Kini, R.; Subramanian, M.; Ohta, A.; Sitkovsky, M. In Vivo T cell activation in lymphoid tissues is inhibited in the oxygen-poor microenvironment. Front. Immunol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Zhuang, X.; Pedroza-Pacheco, I.; Nawroth, I.; Kliszczak, A.E.; Magri, A.; Paes, W.; Rubio, C.O.; Yang, H.; Ashcroft, M.; Mole, D.; et al. Hypoxic microenvironment shapes HIV-1 replication and latency. Commun. Biol. 2020, 3, 376. [Google Scholar] [CrossRef]
- Charles, S.; Ammosova, T.; Cardenas, J.; Foster, A.; Rotimi, J.; Jerebtsova, M.; Ayodeji, A.A.; Niu, X.; Ray, P.E.; Gordeuk, V.R.; et al. Regulation of HIV-1 transcription at 3% versus 21% oxygen concentration. J. Cell Physiol. 2009, 221, 469–479. [Google Scholar] [CrossRef]
- Duette, G.; Pereyra Gerber, P.; Rubione, J.; Perez, P.S.; Landay, A.L.; Crowe, S.M.; Liao, Z.; Witwer, K.W.; Holgado, M.P.; Salido, J.; et al. Induction of HIF-1α by HIV-1 Infection in CD4+ T Cells Promotes Viral Replication and Drives Extracellular Vesicle-Mediated Inflammation. mBio 2018, 9. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Mukerjee, R.; Fan, S.; Del Valle, L.; Michiels, C.; Sweet, T.; Rom, I.; Khalili, K.; Rappaport, J.; Amini, S.; et al. Activation of the oxidative stress pathway by HIV-1 Vpr leads to induction of hypoxia-inducible factor 1alpha expression. J. Biol. Chem. 2009, 284, 11364–11373. [Google Scholar] [CrossRef] [PubMed]
- Barrero, C.A.; Datta, P.K.; Sen, S.; Deshmane, S.; Amini, S.; Khalili, K.; Merali, S. HIV-1 Vpr modulates macrophage metabolic pathways: A SILAC-based quantitative analysis. PLoS ONE 2013, 8, e68376. [Google Scholar] [CrossRef]
- Korgaonkar, S.N.; Feng, X.; Ross, M.D.; Lu, T.C.; D’Agati, V.; Iyengar, R.; Klotman, P.E.; He, J.C. HIV-1 upregulates VEGF in podocytes. J. Am. Soc. Nephrol. 2008, 19, 877–883. [Google Scholar] [CrossRef]
- Kulkarni, A.; Mateus, M.; Thinnes, C.C.; McCullagh, J.S.; Schofield, C.J.; Taylor, G.P.; Bangham, C.R.M. Glucose Metabolism and Oxygen Availability Govern Reactivation of the Latent Human Retrovirus HTLV-1. Cell Chem. Biol. 2017, 24, 1377–1387.e3. [Google Scholar] [CrossRef] [PubMed]
- Mühleisen, A.; Giaisi, M.; Köhler, R.; Krammer, P.H.; Li-Weber, M. Tax contributes apoptosis resistance to HTLV-1-infected T cells via suppression of Bid and Bim expression. Cell Death Dis. 2014, 5, e1575. [Google Scholar] [CrossRef]
- Wakisaka, N.; Kondo, S.; Yoshizaki, T.; Murono, S.; Furukawa, M.; Pagano, J.S. Epstein-Barr virus latent membrane protein 1 induces synthesis of hypoxia-inducible factor 1 alpha. Mol. Cell Biol. 2004, 24, 5223–5234. [Google Scholar] [CrossRef] [PubMed]
- Kraus, R.J.; Yu, X.; Cordes, B.-l.A.; Sathiamoorthi, S.; Iempridee, T.; Nawandar, D.M.; Ma, S.; Romero-Masters, J.C.; McChesney, K.G.; Lin, Z.; et al. Hypoxia-inducible factor-1α plays roles in Epstein-Barr virus’s natural life cycle and tumorigenesis by inducing lytic infection through direct binding to the immediate-early BZLF1 gene promoter. PLoS Pathog. 2017, 13, e1006404. [Google Scholar] [CrossRef]
- Kraus, R.J.; Cordes, B.-l.A.; Sathiamoorthi, S.; Patel, P.; Yuan, X.; Iempridee, T.; Yu, X.; Lee, D.L.; Lambert, P.F.; Mertz, J.E. Reactivation of Epstein-Barr Virus by HIF-1α Requires p53. J. Virol. 2020, 94, e00722-20. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.K.; Dawson, C.W.; Young, L.S.; Ko, C.W.; Hau, P.M.; Lo, K.W. Activation of the FGFR1 signalling pathway by the Epstein-Barr virus-encoded LMP1 promotes aerobic glycolysis and transformation of human nasopharyngeal epithelial cells. J. Pathol. 2015, 237, 238–248. [Google Scholar] [CrossRef]
- Wakisaka, N.; Pagano, J.S. Epstein-Barr virus induces invasion and metastasis factors. Anticancer Res. 2003, 23, 2133–2138. [Google Scholar] [PubMed]
- Kondo, S.; Seo, S.Y.; Yoshizaki, T.; Wakisaka, N.; Furukawa, M.; Joab, I.; Jang, K.L.; Pagano, J.S. EBV latent membrane protein 1 up-regulates hypoxia-inducible factor 1alpha through Siah1-mediated down-regulation of prolyl hydroxylases 1 and 3 in nasopharyngeal epithelial cells. Cancer Res. 2006, 66, 9870–9877. [Google Scholar] [CrossRef] [PubMed]
- Sung, W.W.; Chen, P.R.; Liao, M.H.; Lee, J.W. Enhanced aerobic glycolysis of nasopharyngeal carcinoma cells by Epstein-Barr virus latent membrane protein 1. Exp. Cell Res. 2017, 359, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Ma, B.B.; Hui, E.P.; Wong, S.C.; Mo, F.K.; Leung, S.F.; Kam, M.K.; Chan, A.T. Cyclooxygenase-2 expression in advanced nasopharyngeal carcinoma—A prognostic evaluation and correlation with hypoxia inducible factor 1alpha and vascular endothelial growth factor. Oral. Oncol. 2007, 43, 373–378. [Google Scholar] [CrossRef]
- Yang, L.; Liu, L.; Xu, Z.; Liao, W.; Feng, D.; Dong, X.; Xu, S.; Xiao, L.; Lu, J.; Luo, X.; et al. EBV-LMP1 targeted DNAzyme enhances radiosensitivity by inhibiting tumor angiogenesis via the JNKs/HIF-1 pathway in nasopharyngeal carcinoma. Oncotarget 2015, 6, 5804–5817. [Google Scholar] [CrossRef] [PubMed]
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef]
- Jessie, K.; Fong, M.Y.; Devi, S.; Lam, S.K.; Wong, K.T. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J. Infect. Dis. 2004, 189, 1411–1418. [Google Scholar] [CrossRef]
- Kyle, J.L.; Beatty, P.R.; Harris, E. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J. Infect. Dis. 2007, 195, 1808–1817. [Google Scholar] [CrossRef]
- Pillai, A.B.; Muthuraman, K.R.; Mariappan, V.; Belur, S.S.; Lokesh, S.; Rajendiran, S. Oxidative stress response in the pathogenesis of dengue virus virulence, disease prognosis and therapeutics: An update. Arch. Virol. 2019, 164, 2895–2908. [Google Scholar] [CrossRef]
- Gan, E.S.; Cheong, W.F.; Chan, K.R.; Ong, E.Z.; Chai, X.; Tan, H.C.; Ghosh, S.; Wenk, M.R.; Ooi, E.E. Hypoxia enhances antibody-dependent dengue virus infection. EMBO J. 2017, 36, 1348–1363. [Google Scholar] [CrossRef]
- Frakolaki, E.; Kaimou, P.; Moraiti, M.; Kalliampakou, K.I.; Karampetsou, K.; Dotsika, E.; Liakos, P.; Vassilacopoulou, D.; Mavromara, P.; Bartenschlager, R.; et al. The Role of Tissue Oxygen Tension in Dengue Virus Replication. Cells 2018, 7, 241. [Google Scholar] [CrossRef]
- Olagnier, D.; Peri, S.; Steel, C.; van Montfoort, N.; Chiang, C.; Beljanski, V.; Slifker, M.; He, Z.; Nichols, C.N.; Lin, R.; et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog. 2014, 10, e1004566. [Google Scholar] [CrossRef]
- Le, Q.T.; Chen, E.; Salim, A.; Cao, H.; Kong, C.S.; Whyte, R.; Donington, J.; Cannon, W.; Wakelee, H.; Tibshirani, R.; et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin. Cancer Res. 2006, 12, 1507–1514. [Google Scholar] [CrossRef]
- Shimoda, L.A.; Semenza, G.L. HIF and the lung: Role of hypoxia-inducible factors in pulmonary development and disease. Am. J. Respir. Crit. Care Med. 2011, 183, 152–156. [Google Scholar] [CrossRef]
- van Riel, D.; den Bakker, M.A.; Leijten, L.M.; Chutinimitkul, S.; Munster, V.J.; de Wit, E.; Rimmelzwaan, G.F.; Fouchier, R.A.; Osterhaus, A.D.; Kuiken, T. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am. J. Pathol. 2010, 176, 1614–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, J.; Cheng, L.; Xu, K.; Yang, Y.; Su, X. Deficiency of HIF-1α enhances influenza A virus replication by promoting autophagy in alveolar type II epithelial cells. Emerg. Microbes Infect. 2020, 9, 691–706. [Google Scholar] [CrossRef]
- Thaker, S.K.; Ch’ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef]
- Ritter, J.B.; Wahl, A.S.; Freund, S.; Genzel, Y.; Reichl, U. Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC Syst. Biol. 2010, 4, 61. [Google Scholar] [CrossRef]
- Smallwood, H.S.; Duan, S.; Morfouace, M.; Rezinciuc, S.; Shulkin, B.L.; Shelat, A.; Zink, E.E.; Milasta, S.; Bajracharya, R.; Oluwaseum, A.J.; et al. Targeting Metabolic Reprogramming by Influenza Infection for Therapeutic Intervention. Cell Rep. 2017, 19, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, G.; Xu, Z.G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Koul, N.; Dixit, D.; Sharma, V.; Sen, E. IGF-1 induced HIF-1α-TLR9 cross talk regulates inflammatory responses in glioma. Cell. Signal. 2011, 23, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Zampell, J.C.; Yan, A.; Avraham, T.; Daluvoy, S.; Weitman, E.S.; Mehrara, B.J. HIF-1α coordinates lymphangiogenesis during wound healing and in response to inflammation. FASEB J. 2012, 26, 1027–1039. [Google Scholar] [CrossRef]
- Xi, Y.; Kim, T.; Brumwell, A.N.; Driver, I.H.; Wei, Y.; Tan, V.; Jackson, J.R.; Xu, J.; Lee, D.K.; Gotts, J.E.; et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 2017, 19, 904–914. [Google Scholar] [CrossRef]
- Yee, M.; Buczynski, B.W.; Lawrence, B.P.; O’Reilly, M.A. Neonatal hyperoxia increases sensitivity of adult mice to bleomycin-induced lung fibrosis. Am. J. Respir. Cell Mol. Biol. 2013, 48, 258–266. [Google Scholar] [CrossRef]
- Buczynski, B.W.; Yee, M.; Martin, K.C.; Lawrence, B.P.; O’Reilly, M.A. Neonatal hyperoxia alters the host response to influenza A virus infection in adult mice through multiple pathways. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 305, L282–L290. [Google Scholar] [CrossRef]
- Kumar, V.H.S.; Wang, H.; Nielsen, L. Adaptive immune responses are altered in adult mice following neonatal hyperoxia. Physiol. Rep. 2018, 6. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, W.; Han, P.; Zhang, J.; Zhu, Y.; Meng, X.; Zhang, J.; Hu, Y.; Yi, Z.; Wang, R. Influenza A virus (H1N1) triggers a hypoxic response by stabilizing hypoxia-inducible factor-1α via inhibition of proteasome. Virology 2019, 530, 51–58. [Google Scholar] [CrossRef]
- Mühlbauer, D.; Dzieciolowski, J.; Hardt, M.; Hocke, A.; Schierhorn, K.L.; Mostafa, A.; Müller, C.; Wisskirchen, C.; Herold, S.; Wolff, T.; et al. Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes. J. Virol. 2015, 89, 6009–6021. [Google Scholar] [CrossRef]
- Huo, C.; Wu, H.; Xiao, J.; Meng, D.; Zou, S.; Wang, M.; Qi, P.; Tian, H.; Hu, Y. Genomic and Bioinformatic Characterization of Mouse Mast Cells (P815) upon Different Influenza A Virus (H1N1, H5N1, and H7N2) Infections. Front. Genet. 2019, 10, 595. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yin, G.; Ma, Y.; Xu, K.; Liu, J.; Li, J. The critical role of mast cell-derived hypoxia-inducible factor-1α in regulating mast cell function. J. Pharm. Pharmacol. 2016, 68, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., 3rd; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446.e14. [Google Scholar] [CrossRef]
- Guan, W.J.; Liang, W.H.; Zhao, Y.; Liang, H.R.; Chen, Z.S.; Li, Y.M.; Liu, X.Q.; Chen, R.C.; Tang, C.L.; Wang, T.; et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: A nationwide analysis. Eur. Respir. J. 2020, 55. [Google Scholar] [CrossRef]
- Marchetti, M. COVID-19-driven endothelial damage: Complement, HIF-1, and ABL2 are potential pathways of damage and targets for cure. Ann. Hematol. 2020, 99, 1701–1707. [Google Scholar] [CrossRef]
- Gavriatopoulou, M.; Korompoki, E.; Fotiou, D.; Ntanasis-Stathopoulos, I.; Psaltopoulou, T.; Kastritis, E.; Terpos, E.; Dimopoulos, M.A. Organ-specific manifestations of COVID-19 infection. Clin. Exp. Med. 2020. [Google Scholar] [CrossRef]
- Trottein, F.; Sokol, H. Potential Causes and Consequences of Gastrointestinal Disorders during a SARS-CoV-2 Infection. Cell Rep. 2020, 32, 107915. [Google Scholar] [CrossRef]
- Marshall, M. How COVID-19 can damage the brain. Nature 2020, 585, 342–343. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, Y.; Zhao, M.; Liu, C.; Zhou, L.; Shen, S.; Liao, S.; Yang, K.; Li, Q.; Wan, H. Role of HIF-1alpha in the regulation ACE and ACE2 expression in hypoxic human pulmonary artery smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 297, L631–L640. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Wollenzien, H.; Leclerc, E.; Jarajapu, Y.P. Hypoxic regulation of angiotensin-converting enzyme 2 and Mas receptor in human CD34+ cells. J. Cell. Physiol. 2019, 234, 20420–20431. [Google Scholar] [CrossRef] [PubMed]
- Wing, P.A.; Keeley, T.P.; Zhuang, X.; Lee, J.Y.; Prange-Barczynska, M.; Tsukuda, S.; Morgan, S.B.; Argles, I.L.A.; Kurlekar, S.; Noerenberg, M.; et al. Hypoxic and pharmacological activation of HIF inhibits SARS-CoV-2 infection of lung epithelial cells. bioRxiv 2020, 13, 494–499. [Google Scholar]
- Yee, M.; Cohen, E.D.; Haak, J.; Dylag, A.M.; O’Reilly, M.A. Neonatal hyperoxia enhances age-dependent expression of SARS-CoV-2 receptors in mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; de Brito Monteiro, L.; Fabiano de Souza, G.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; Oliveira de Biagi Junior, C.A.; Crunfli, F.; et al. Title: Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis dependent axis. Cell Metab. 2020. [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ní Choileáin, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care. Med. 2020. [Google Scholar] [CrossRef]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; Weizman, O.-E.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Appelberg, S.; Gupta, S.; Svensson Akusjärvi, S.; Ambikan, A.T.; Mikaeloff, F.; Saccon, E.; Végvári, Á.; Benfeitas, R.; Sperk, M.; Ståhlberg, M.; et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes Infect. 2020, 9, 1748–1760. [Google Scholar] [CrossRef]
- Chen, R.; Forsyth, N. Editorial: The Development of New Classes of Hypoxia Mimetic Agents for Clinical Use. Front. Cell. Dev. Biol. 2019, 7, 120. [Google Scholar] [CrossRef]
- Phase II Multicentric Study of Digoxin Per os in Classic or Endemic Kaposi’s Sarcoma. Available online: https://ClinicalTrials.gov/show/NCT02212639 (accessed on 4 September 2020).
- Krasner, C.; Birrer, M.; Peters, C.; Jayaraman, L.; Eliasof, S.; Tellez, A.; Downing, W.; Senderowicz, A. Abstract CT090: Phase II trial of the NDC CRLX101 in combination with bevacizumab in patients with platinum-resistant ovarian cancer (PROC). Cancer Res. 2016, 76, CT090. [Google Scholar] [CrossRef]
- Pham, E.; Birrer, M.J.; Eliasof, S.; Garmey, E.G.; Lazarus, D.; Lee, C.R.; Man, S.; Matulonis, U.A.; Peters, C.G.; Xu, P.; et al. Translational impact of nanoparticle-drug conjugate CRLX101 with or without bevacizumab in advanced ovarian cancer. Clin. Cancer Res. 2015, 21, 808–818. [Google Scholar] [CrossRef]
- Voss, M.H.; Hussain, A.; Vogelzang, N.; Lee, J.L.; Keam, B.; Rha, S.Y.; Vaishampayan, U.; Harris, W.B.; Richey, S.; Randall, J.M.; et al. A randomized phase II trial of CRLX101 in combination with bevacizumab versus standard of care in patients with advanced renal cell carcinoma. Ann. Oncol. 2017, 28, 2754–2760. [Google Scholar] [CrossRef] [PubMed]
- Sanoff, H.K.; Moon, D.H.; Moore, D.T.; Boles, J.; Bui, C.; Blackstock, W.; O’Neil, B.H.; Subramaniam, S.; McRee, A.J.; Carlson, C.; et al. Phase I/II trial of nano-camptothecin CRLX101 with capecitabine and radiotherapy as neoadjuvant treatment for locally advanced rectal cancer. Nanomed. Nanotechnol. Biol. Med. 2019, 18, 189–195. [Google Scholar] [CrossRef]
- Renfrow, J.J.; Soike, M.H.; Debinski, W.; Ramkissoon, S.H.; Mott, R.T.; Frenkel, M.B.; Sarkaria, J.N.; Lesser, G.J.; Strowd, R.E. Hypoxia-inducible factor 2α: A novel target in gliomas. Future Med. Chem. 2018, 10, 2227–2236. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, Q.; Zhang, X. Allosteric inhibition of HIF-2α as a novel therapy for clear cell renal cell carcinoma. Drug Discov. Today 2019, 24, 2332–2340. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, K.; Rizzi, J.P.; Huang, H.; Grina, J.A.; Schlachter, S.T.; Wang, B.; Wehn, P.M.; Yang, H.; Dixon, D.D.; et al. 3-[(1S,2S,3R)-2,3-Difluoro-1-hydroxy-7-methylsulfonylindan-4-yl]oxy-5-fluorobenzonitrile (PT2977), a Hypoxia-Inducible Factor 2α (HIF-2α) Inhibitor for the Treatment of Clear Cell Renal Cell Carcinoma. J. Med. Chem. 2019, 62, 6876–6893. [Google Scholar] [CrossRef] [PubMed]
- A Trial of PT2977 Tablets In Patients with Advanced Solid Tumors. Available online: https://ClinicalTrials.gov/show/NCT02974738 (accessed on 4 September 2020).
- HIF-2 Alpha Inhibitor PT2385 in Treating Patients with Recurrent Glioblastoma. Available online: https://ClinicalTrials.gov/show/NCT03216499 (accessed on 4 September 2020).
- Strowd, R.; Ellingson, B.; Wen, P.; Ahluwalia, M.; Piotrowski, A.; Desai, A.; Clarke, J.; Lieberman, F.; Desideri, S.; Nabors, L.B.; et al. ACTR-15. Safety and preliminary activity of PT2385, a first-in-class HIF2-alpha inhibitor, planned interim analysis of an open label, single-arm phase II study in patients with recurrent glioblastoma. Neuro-Oncology 2018, 20, vi14. [Google Scholar] [CrossRef]
- Gilmore, S.A.; Snyder, C.A.; Dick, R.; Matles, M.; Tam, D.; Tay, C.H.; Farand, J.; Paoli, E.; Delaney, W.E.; Feierbach, B.; et al. THU-171 - In Vivo pharmacodynamics of GS-5801, a liver targeted prodrug of a lysine demethylase 5 inhibitor with antiviral activity against hepatitis B virus. J. Hepatol. 2017, 66, S263. [Google Scholar] [CrossRef]
- Wu, L.; Cao, J.; Cai, W.L.; Lang, S.M.; Horton, J.R.; Jansen, D.J.; Liu, Z.Z.; Chen, J.F.; Zhang, M.; Mott, B.T.; et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 2018, 16, e2006134. [Google Scholar] [CrossRef]
- Post, D.E.; Van Meir, E.G. A novel hypoxia-inducible factor (HIF) activated oncolytic adenovirus for cancer therapy. Oncogene 2003, 22, 2065–2072. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.K.; Seong, Y.R.; Lee, Y.H.; Kim, M.J.; Hwang, K.S.; Yoo, J.; Choi, S.; Jung, C.R.; Im, D.S. Oncolytic effects of adenovirus mutant capable of replicating in hypoxic and normoxic regions of solid tumor. Mol. Ther. 2004, 10, 938–949. [Google Scholar] [CrossRef]
- Mazzon, M.; Peters, N.E.; Loenarz, C.; Krysztofinska, E.M.; Ember, S.W.; Ferguson, B.J.; Smith, G.L. A mechanism for induction of a hypoxic response by vaccinia virus. Proc. Natl. Acad. Sci. USA 2013, 110, 12444–12449. [Google Scholar] [CrossRef] [PubMed]
- Mazzon, M.; Castro, C.; Roberts, L.D.; Griffin, J.L.; Smith, G.L. A role for vaccinia virus protein C16 in reprogramming cellular energy metabolism. J. Gen. Virol. 2015, 96, 395–407. [Google Scholar] [CrossRef]
- Cho, I.R.; Koh, S.S.; Min, H.J.; Park, E.H.; Ratakorn, S.; Jhun, B.H.; Jeong, S.H.; Yoo, Y.H.; Youn, H.D.; Johnston, R.N.; et al. Down-regulation of HIF-1alpha by oncolytic reovirus infection independently of VHL and p53. Cancer Gene Ther. 2010, 17, 365–372. [Google Scholar] [CrossRef]
- Gupta-Saraf, P.; Miller, C.L. HIF-1α downregulation and apoptosis in hypoxic prostate tumor cells infected with oncolytic mammalian orthoreovirus. Oncotarget 2014, 5, 561–574. [Google Scholar] [CrossRef]
- Hotani, T.; Tachibana, M.; Mizuguchi, H.; Sakurai, F. Reovirus double-stranded RNA genomes and polyI:C induce down-regulation of hypoxia-inducible factor 1α. Biochem. Biophys. Res. Commun. 2015, 460, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Hotani, T.; Mizuguchi, H.; Sakurai, F. Systemically Administered Reovirus-Induced Downregulation of Hypoxia Inducible Factor-1α in Subcutaneous Tumors. Mol. Ther. Oncolytics 2018, 12, 162–172. [Google Scholar] [CrossRef]
- Gong, J.; Sachdev, E.; Mita, A.C.; Mita, M.M. Clinical development of reovirus for cancer therapy: An oncolytic virus with immune-mediated antitumor activity. World J. Methodol. 2016, 6, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Cheng, K.; Chandel, N.; Lederman, R.; Jhaveri, A.; Husain, M.; Malhotra, A.; Singhal, P.C. High glucose enhances HIV entry into T cells through upregulation of CXCR4. J. Leukoc. Biol. 2013, 94, 769–777. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).