“FastCheckFLI PPR-like”—A Molecular Tool for the Fast Genome Detection of PPRV and Differential Diagnostic Pathogens
Abstract
1. Introduction
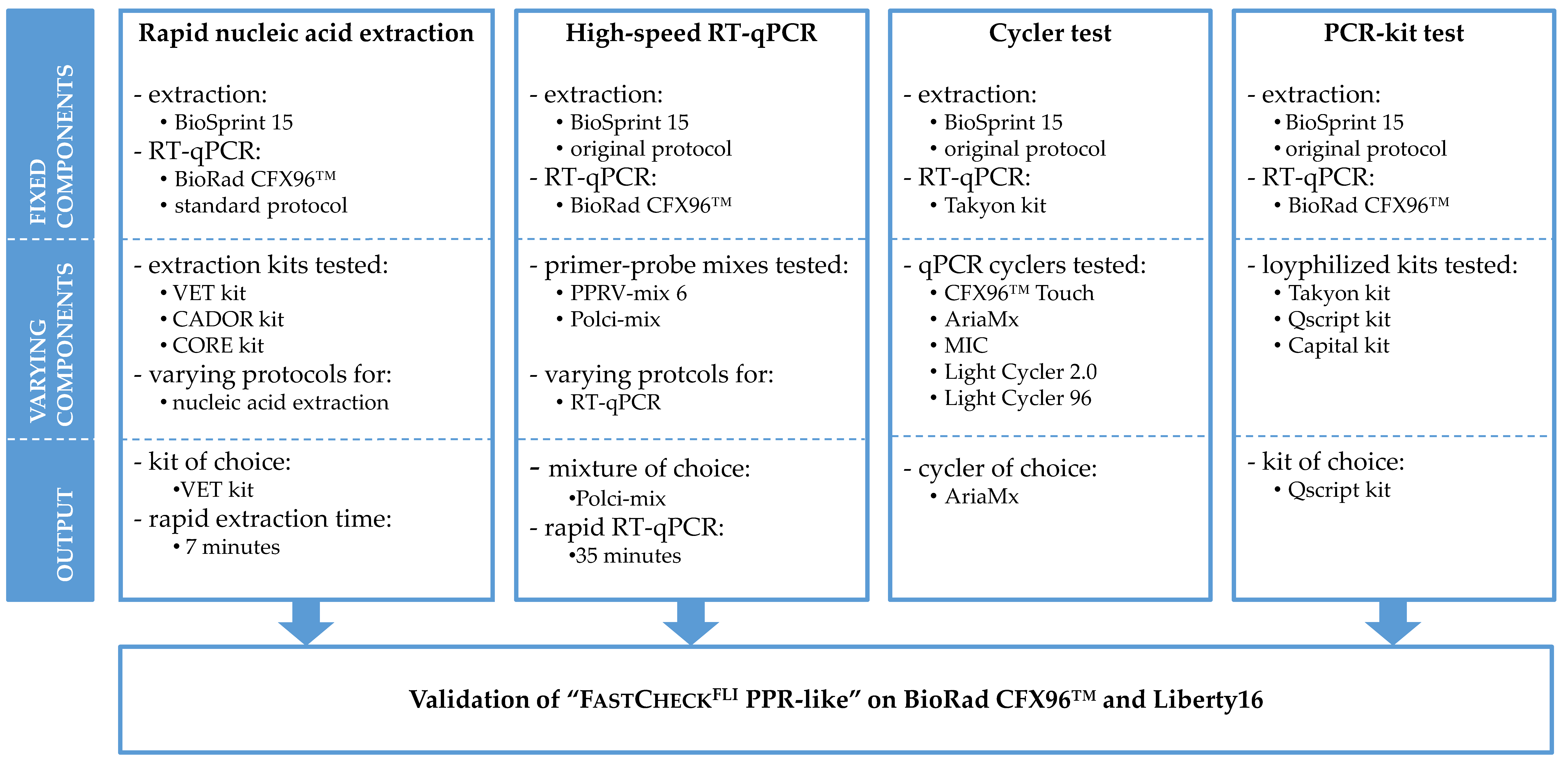
2. Materials and Methods
2.1. Rapid Nucleic Acid Extraction
2.2. High-Speed RT-qPCR
2.3. Device Test of Different qPCR Cyclers
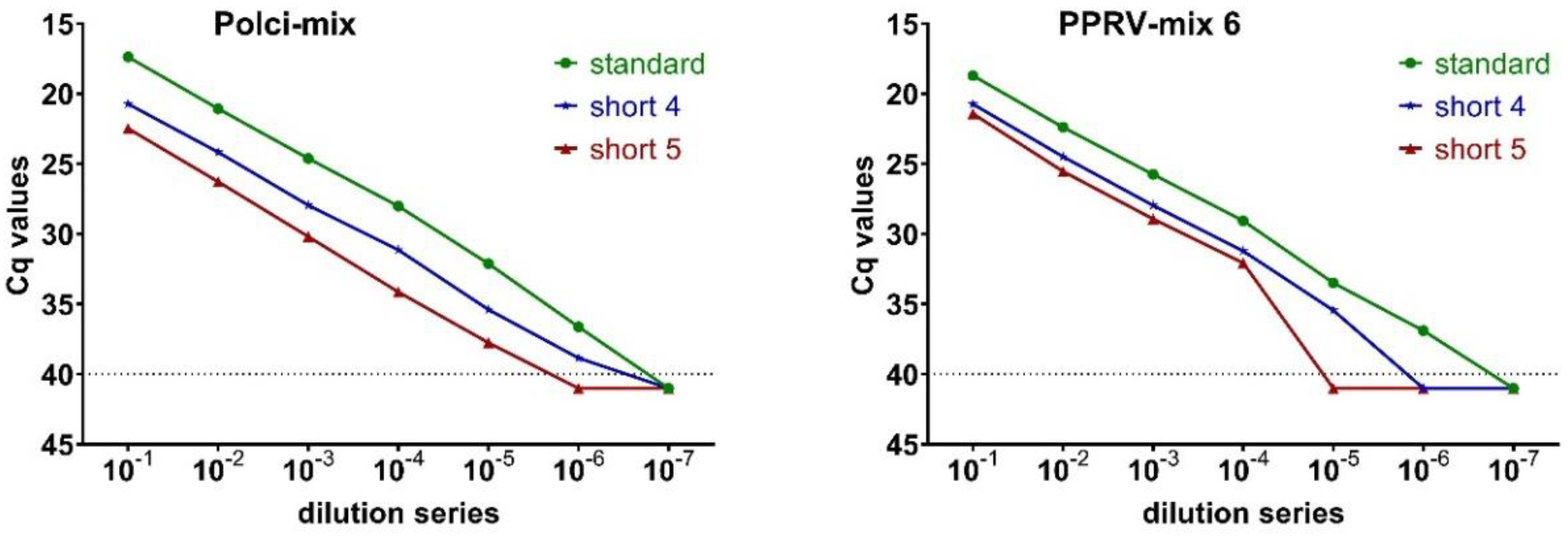
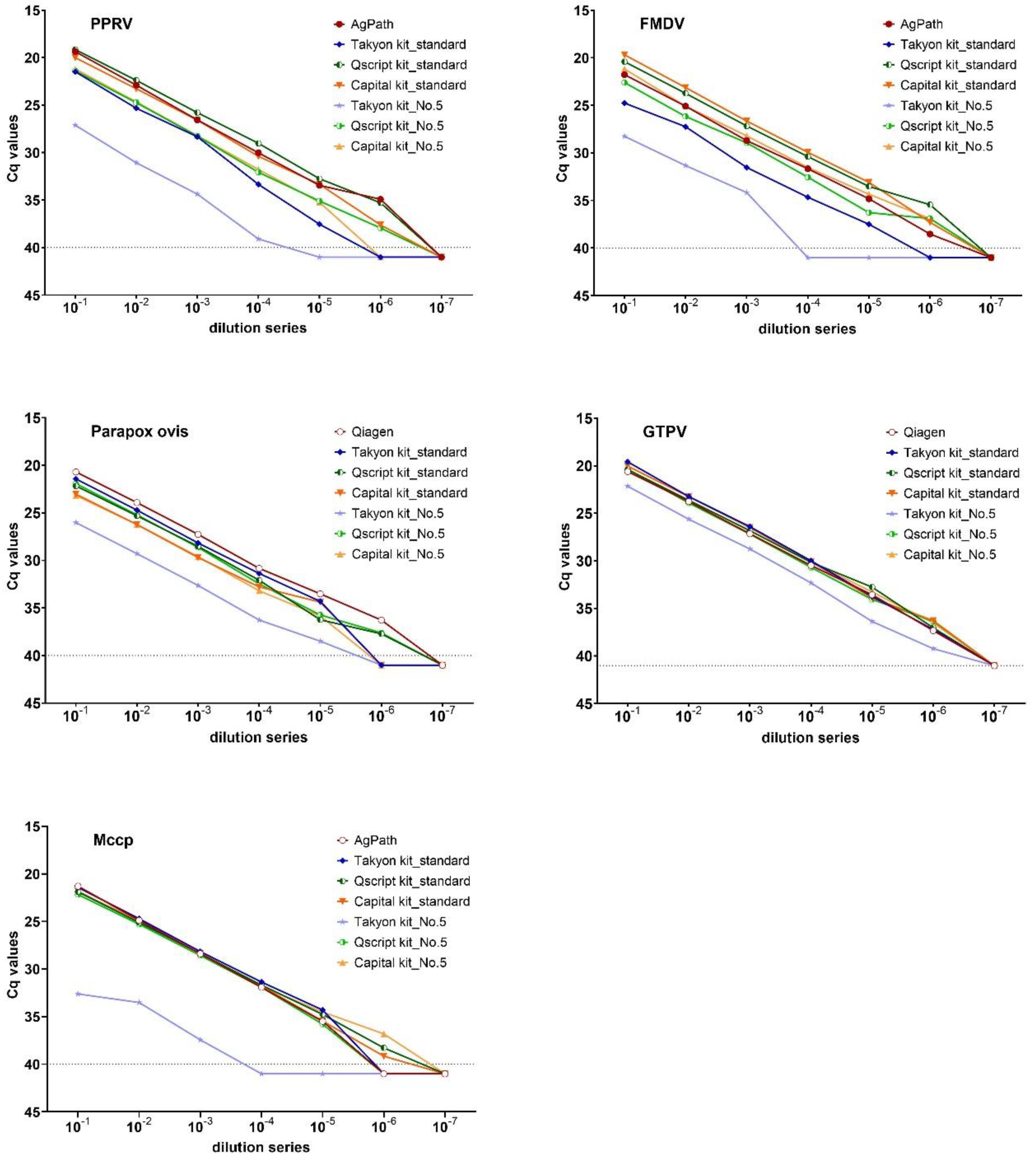
2.4. Validation of Three Lyophilized Kits
2.5. Validation of the “FastCheckFLI PPR-Like” System
3. Results
3.1. Speed-Optimized Rapid Extraction Protocols
3.2. PPRV-Specific High-Speed RT-qPCR
3.3. Assessment of the Fast Cycling Features of Five qPCR Cyclers
3.4. Evaluation of Lyophilized Kits for POC Testing
3.5. Validation of the “FastCheckFLI PPR-Like” Workflow
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Munir, M. Role of wild small ruminants in the epidemiology of peste des petits ruminants. Transbound Emerg. Dis. 2014, 61, 411–424. [Google Scholar] [CrossRef]
- Wensman, J.J.; Abubakar, M.; Shabbir, M.Z.; Rossiter, P. Peste des petits ruminants in wild ungulates. Trop. Anim. Health Prod. 2018, 50, 1815–1819. [Google Scholar] [CrossRef]
- Schulz, C.; Fast, C.; Schlottau, K.; Hoffmann, B.; Beer, M. Neglected Hosts of Small Ruminant Morbillivirus. Emerg. Infect. Dis. 2018, 24, 2334–2337. [Google Scholar] [CrossRef] [PubMed]
- Schulz, C.; Fast, C.; Wernery, U.; Kinne, J.; Joseph, S.; Schlottau, K.; Jenckel, M.; Hoper, D.; Patteril, N.A.G.; Syriac, G.; et al. Camelids and Cattle Are Dead-End Hosts for Peste-des-Petits-Ruminants Virus. Viruses 2019, 11, 1133. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, G.K.; Bao, Y.; Basler, C.F.; Bavari, S.; Beer, M.; Bejerman, N.; Blasdell, K.R.; Bochnowski, A.; Briese, T.; Bukreyev, A.; et al. Taxonomy of the order Mononegavirales: Update 2017. Arch. Virol. 2017, 162, 2493–2504. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Maherchandani, S.; Kashyap, S.K.; Singh, S.V.; Sharma, S.; Chaubey, K.K.; Ly, H. Peste des petits ruminants virus infection of small ruminants: A comprehensive review. Viruses 2014, 6, 2287–2327. [Google Scholar] [CrossRef]
- Shaila, M.S.; Shamaki, D.; Forsyth, M.A.; Diallo, A.; Goatley, L.; Kitching, R.P.; Barrett, T. Geographic distribution and epidemiology of peste des petits ruminants virus. Virus Res. 1996, 43, 149–153. [Google Scholar] [CrossRef]
- Dhar, P.; Sreenivasa, B.P.; Barrett, T.; Corteyn, M.; Singh, R.P.; Bandyopadhyay, S.K. Recent epidemiology of peste des petits ruminants virus (PPRV). Vet. Microbiol. 2002, 88, 153–159. [Google Scholar] [CrossRef]
- Parida, S.; Muniraju, M.; Mahapatra, M.; Muthuchelvan, D.; Buczkowski, H.; Banyard, A.C. Peste des petits ruminants. Vet. Microbiol. 2015, 181, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Spyrou, V.; Valiakos, G. Orf virus infection in sheep or goats. Vet. Microbiol. 2015, 181, 178–182. [Google Scholar] [CrossRef]
- Kumar, N.; Barua, S.; Riyesh, T.; Chaubey, K.K.; Rawat, K.D.; Khandelwal, N.; Mishra, A.K.; Sharma, N.; Chandel, S.S.; Sharma, S.; et al. Complexities in Isolation and Purification of Multiple Viruses from Mixed Viral Infections: Viral Interference, Persistence and Exclusion. PLoS ONE 2016, 11, e0156110. [Google Scholar] [CrossRef] [PubMed]
- Adedeji, A.J.; Dashe, Y.; Akanbi, O.B.; Woma, T.Y.; Jambol, A.R.; Adole, J.A.; Bolajoko, M.B.; Chima, N.; Asala, O.; Tekki, I.S.; et al. Co-infection of peste des petits ruminants and goatpox in a mixed flock of sheep and goats in Kanam, North Central Nigeria. Vet. Med. Sci. 2019, 5, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, P.; Balamurugan, V.; Sen, A.; Sarkar, J.; Sahay, B.; Rajak, K.K.; Hosamani, M.; Yadav, M.P.; Singh, R.K. Mixed infection of peste des petits ruminants and orf on a goat farm in Shahjahanpur, India. Vet. Rec. 2007, 160, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Jamal, S.M.; Belsham, G.J. Foot-and-mouth disease: Past, present and future. Vet. Res. 2013, 44, 116. [Google Scholar] [CrossRef]
- Alexandersen, S.; Zhang, Z.; Donaldson, A.I.; Garland, A.J. The pathogenesis and diagnosis of foot-and-mouth disease. J. Comp. Pathol. 2003, 129, 1–36. [Google Scholar] [CrossRef]
- Tuppurainen, E.S.M.; Venter, E.H.; Shisler, J.L.; Gari, G.; Mekonnen, G.A.; Juleff, N.; Lyons, N.A.; De Clercq, K.; Upton, C.; Bowden, T.R.; et al. Review: Capripoxvirus Diseases: Current Status and Opportunities for Control. Transbound Emerg. Dis. 2017, 64, 729–745. [Google Scholar] [CrossRef]
- Babiuk, S.; Bowden, T.R.; Boyle, D.B.; Wallace, D.B.; Kitching, R.P. Capripoxviruses: An emerging worldwide threat to sheep, goats and cattle. Transbound Emerg. Dis. 2008, 55, 263–272. [Google Scholar] [CrossRef]
- Rao, T.V.; Bandyopadhyay, S.K. A comprehensive review of goat pox and sheep pox and their diagnosis. Anim. Health Res. Rev. 2000, 1, 127–136. [Google Scholar] [CrossRef]
- Iqbal Yatoo, M.; Raffiq Parray, O.; Tauseef Bashir, S.; Ahmed Bhat, R.; Gopalakrishnan, A.; Karthik, K.; Dhama, K.; Vir Singh, S. Contagious caprine pleuropneumonia—A comprehensive review. Vet. Q. 2019, 39, 1–25. [Google Scholar] [CrossRef]
- Nicholas, R.; Churchward, C. Contagious caprine pleuropneumonia: New aspects of an old disease. Transbound Emerg. Dis. 2012, 59, 189–196. [Google Scholar] [CrossRef]
- Arif, A.; Schulz, J.; Thiaucourt, F.; Taha, A.; Hammer, S. Contagious caprine pleuropneumonia outbreak in captive wild ungulates at Al Wabra Wildlife Preservation, State of Qatar. J. Zoo Wildl. Med. 2007, 38, 93–96. [Google Scholar] [CrossRef]
- Banyard, A.C.; Parida, S.; Batten, C.; Oura, C.; Kwiatek, O.; Libeau, G. Global distribution of peste des petits ruminants virus and prospects for improved diagnosis and control. J. Gen. Virol. 2010, 91, 2885–2897. [Google Scholar] [CrossRef] [PubMed]
- Albina, E.; Kwiatek, O.; Minet, C.; Lancelot, R.; Servan de Almeida, R.; Libeau, G. Peste des Petits Ruminants, the next eradicated animal disease? Vet. Microbiol. 2013, 165, 38–44. [Google Scholar] [CrossRef] [PubMed]
- FAO. Mapping Achievements towards the Eradication of Peste des Petits Ruminant (PPR); FAO, Ed.; FAO: Rome, Italy, 2018; p. 1. Available online: http://www.fao.org/3/CA1373EN/ca1373en.pdf (accessed on 16 October 2019).
- Li, Y.; Li, L.; Fan, X.; Zou, Y.; Zhang, Y.; Wang, Q.; Sun, C.; Pan, S.; Wu, X.; Wang, Z. Development of real-time reverse transcription recombinase polymerase amplification (RPA) for rapid detection of peste des petits ruminants virus in clinical samples and its comparison with real-time PCR test. Sci. Rep. 2018, 8, 17760. [Google Scholar] [CrossRef] [PubMed]
- Howson, E.L.A.; Soldan, A.; Webster, K.; Beer, M.; Zientara, S.; Belak, S.; Sanchez-Vizcaino, J.M.; Van Borm, S.; King, D.P.; Fowler, V.L. Technological advances in veterinary diagnostics: Opportunities to deploy rapid decentralised tests to detect pathogens affecting livestock. Rev. Sci. Tech. 2017, 36, 479–498. [Google Scholar] [CrossRef]
- Ashraf, W.; Unger, H.; Haris, S.; Mobeen, A.; Farooq, M.; Asif, M.; Khan, Q.M. Genetic detection of peste des petits ruminants virus under field conditions: A step forward towards disease eradication. BMC Vet. Res. 2017, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Rajko-Nenow, P.; Flannery, J.; Arnold, H.; Howson, E.L.A.; Darpel, K.; Stedman, A.; Corla, A.; Batten, C. A rapid RT-LAMP assay for the detection of all four lineages of Peste des Petits Ruminants Virus. J. Virol. Methods 2019, 274, 113730. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, M.; Howson, E.; Fowler, V.; Batten, C.; Flannery, J.; Selvaraj, M.; Parida, S. Rapid Detection of Peste des Petits Ruminants Virus (PPRV) Nucleic Acid Using a Novel Low-Cost Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Assay for Future Use in Nascent PPR Eradication Programme. Viruses 2019, 11, 699. [Google Scholar] [CrossRef]
- ID.Vet. ID Rapid® PPR Antigen. Available online: https://www.id-vet.com/produit/id-rapid-ppr-antigen/ (accessed on 19 August 2019).
- Baron, J.; Fishbourne, E.; Couacy-Hyman, E.; Abubakar, M.; Jones, B.A.; Frost, L.; Herbert, R.; Chibssa, T.R.; Van’t Klooster, G.; Afzal, M.; et al. Development and testing of a field diagnostic assay for peste des petits ruminants virus. Transbound Emerg. Dis. 2014, 61, 390–396. [Google Scholar] [CrossRef]
- Rozand, C. Paper-based analytical devices for point-of-care infectious disease testing. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 147–156. [Google Scholar] [CrossRef]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 2013, 13, 2210–2251. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.A.; Mahapatra, M.; Chubwa, C.; Clarke, B.; Batten, C.; Hicks, H.; Henstock, M.; Keyyu, J.; Kock, R.; Parida, S. Characterisation of Peste Des Petits Ruminants Disease in Pastoralist Flocks in Ngorongoro District of Northern Tanzania and Bluetongue Virus Co-Infection. Viruses 2020, 12, 389. [Google Scholar] [CrossRef] [PubMed]
- Halecker, S.; Joseph, S.; Mohammed, R.; Wernery, U.; Mettenleiter, T.C.; Beer, M.; Hoffmann, B. Comparative evaluation of different antigen detection methods for the detection of peste des petits ruminants virus. Transbound Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, V.; Sen, A.; Venkatesan, G.; Yadav, V.; Bhanot, V.; Bhanuprakash, V.; Singh, R.K. Application of semi-quantitative M gene-based hydrolysis probe (TaqMan) real-time RT-PCR assay for the detection of peste des petits ruminants virus in the clinical samples for investigation into clinical prevalence of disease. Transbound Emerg. Dis. 2010, 57, 383–395. [Google Scholar] [CrossRef]
- Bao, J.; Li, L.; Wang, Z.; Barrett, T.; Suo, L.; Zhao, W.; Liu, Y.; Liu, C.; Li, J. Development of one-step real-time RT-PCR assay for detection and quantitation of peste des petits ruminants virus. J. Virol. Methods 2008, 148, 232–236. [Google Scholar] [CrossRef]
- Kwiatek, O.; Keita, D.; Gil, P.; Fernandez-Pinero, J.; Jimenez Clavero, M.A.; Albina, E.; Libeau, G. Quantitative one-step real-time RT-PCR for the fast detection of the four genotypes of PPRV. J. Virol. Methods 2010, 165, 168–177. [Google Scholar] [CrossRef]
- Batten, C.A.; Banyard, A.C.; King, D.P.; Henstock, M.R.; Edwards, L.; Sanders, A.; Buczkowski, H.; Oura, C.C.; Barrett, T. A real time RT-PCR assay for the specific detection of Peste des petits ruminants virus. J. Virol. Methods 2011, 171, 401–404. [Google Scholar] [CrossRef]
- Polci, A.; Cosseddu, G.M.; Ancora, M.; Pinoni, C.; El Harrak, M.; Sebhatu, T.T.; Ghebremeskel, E.; Sghaier, S.; Lelli, R.; Monaco, F. Development and Preliminary Evaluation of a New Real-Time RT-PCR Assay for Detection of Peste des Petits Ruminants Virus Genome. Transbound Emerg. Dis. 2015, 62, 332–338. [Google Scholar] [CrossRef]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef]
- Howson, E.L.A.; Armson, B.; Madi, M.; Kasanga, C.J.; Kandusi, S.; Sallu, R.; Chepkwony, E.; Siddle, A.; Martin, P.; Wood, J.; et al. Evaluation of Two Lyophilized Molecular Assays to Rapidly Detect Foot-and-Mouth Disease Virus Directly from Clinical Samples in Field Settings. Transbound Emerg. Dis. 2017, 64, 861–871. [Google Scholar] [CrossRef]
- Righter, D.J.; Rurangirwa, F.R.; Call, D.R.; McElwain, T.F. Development of a bead-based multiplex PCR assay for the simultaneous detection of multiple Mycoplasma species. Vet. Microbiol. 2011, 153, 246–256. [Google Scholar] [CrossRef]
- Wernike, K.; Beer, M.; Hoffmann, B. Rapid detection of foot-and-mouth disease virus, influenza A virus and classical swine fever virus by high-speed real-time RT-PCR. J. Virol. Methods 2013, 193, 50–54. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nitsche, A.; Buttner, M.; Wilhelm, S.; Pauli, G.; Meyer, H. Real-time PCR detection of parapoxvirus DNA. Clin. Chem. 2006, 52, 316–319. [Google Scholar] [CrossRef]
- Bowden, T.R.; Babiuk, S.L.; Parkyn, G.R.; Copps, J.S.; Boyle, D.B. Capripoxvirus tissue tropism and shedding: A quantitative study in experimentally infected sheep and goats. Virology 2008, 371, 380–393. [Google Scholar] [CrossRef]
- Dietze, K.; Moritz, T.; Alexandrov, T.; Krstevski, K.; Schlottau, K.; Milovanovic, M.; Hoffmann, D.; Hoffmann, B. Suitability of group-level oral fluid sampling in ruminant populations for lumpy skin disease virus detection. Vet. Microbiol. 2018, 221, 44–48. [Google Scholar] [CrossRef]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, J.F.; Sailleau, C.; Breard, E.; Zientara, S.; De Clercq, K. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J. Virol. Methods 2007, 140, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Hoffmann, B.; Kalthoff, D.; Konig, P.; Beer, M. Development and validation of a triplex real-time PCR assay for the rapid detection and differentiation of wild-type and glycoprotein E-deleted vaccine strains of Bovine herpesvirus type 1. J. Virol. Methods 2011, 174, 77–84. [Google Scholar] [CrossRef]
- Schlottau, K.; Freuling, C.M.; Muller, T.; Beer, M.; Hoffmann, B. Development of molecular confirmation tools for swift and easy rabies diagnostics. Virol. J. 2017, 14, 184. [Google Scholar] [CrossRef]
- Ferris, N.P.; Nordengrahn, A.; Hutchings, G.H.; Reid, S.M.; King, D.P.; Ebert, K.; Paton, D.J.; Kristersson, T.; Brocchi, E.; Grazioli, S.; et al. Development and laboratory validation of a lateral flow device for the detection of foot-and-mouth disease virus in clinical samples. J. Virol. Methods 2009, 155, 10–17. [Google Scholar] [CrossRef]
- Cetre-Sossah, C.; Pedarrieu, A.; Juremalm, M.; Jansen Van Vuren, P.; Brun, A.; Ould El Mamy, A.B.; Heraud, J.M.; Filippone, C.; Ravalohery, J.P.; Chaabihi, H.; et al. Development and validation of a pen side test for Rift Valley fever. PLoS Negl. Trop. Dis. 2019, 13, e0007700. [Google Scholar] [CrossRef]
- Molsa, M.; Koskela, K.A.; Ronkko, E.; Ikonen, N.; Ziegler, T.; Nikkari, S. Detection of influenza A viruses with a portable real-time PCR instrument. J. Virol. Methods 2012, 181, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Bruning-Richardson, A.; Akerblom, L.; Klingeborn, B.; Anderson, J. Improvement and development of rapid chromatographic strip-tests for the diagnosis of rinderpest and peste des petits ruminants viruses. J. Virol. Methods 2011, 174, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Madi, M.; Hamilton, A.; Squirrell, D.; Mioulet, V.; Evans, P.; Lee, M.; King, D.P. Rapid detection of foot-and-mouth disease virus using a field-portable nucleic acid extraction and real-time PCR amplification platform. Vet. J. 2012, 193, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Ferris, N.P.; Nordengrahn, A.; Hutchings, G.H.; Paton, D.J.; Kristersson, T.; Brocchi, E.; Grazioli, S.; Merza, M. Development and laboratory validation of a lateral flow device for the detection of serotype SAT 2 foot-and-mouth disease viruses in clinical samples. J. Virol. Methods 2010, 163, 474–476. [Google Scholar] [CrossRef]
- Jiang, T.; Liang, Z.; Ren, W.; Chen, J.; Zhi, X.; Qi, G.; Yang, Y.; Liu, Z.; Liu, X.; Cai, X. Development and validation of a lateral flow immunoassay using colloidal gold for the identification of serotype-specific foot-and-mouth disease virus O, A and Asia 1. J. Virol. Methods 2011, 171, 74–80. [Google Scholar] [CrossRef]
- Dineva, M.A.; MahiLum-Tapay, L.; Lee, H. Sample preparation: A challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. Analyst 2007, 132, 1193–1199. [Google Scholar] [CrossRef]
- Howson, E.L.A.; Armson, B.; Lyons, N.A.; Chepkwony, E.; Kasanga, C.J.; Kandusi, S.; Ndusilo, N.; Yamazaki, W.; Gizaw, D.; Cleaveland, S.; et al. Direct detection and characterization of foot-and-mouth disease virus in East Africa using a field-ready real-time PCR platform. Transbound Emerg. Dis. 2018, 65, 221–231. [Google Scholar] [CrossRef]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef]
- Aebischer, A.; Beer, M.; Hoffmann, B. Development and validation of rapid magnetic particle based extraction protocols. Virol. J. 2014, 11, 137. [Google Scholar] [CrossRef]
- Holland, C.A.; Kiechle, F.L. Point-of-care molecular diagnostic systems--past, present and future. Curr. Opin. Microbiol. 2005, 8, 504–509. [Google Scholar] [CrossRef]
- Priye, A.; Ugaz, V.M. Smartphone-Enabled Detection Strategies for Portable PCR–Based Diagnostics. In Biosensors and Biodetection: Methods and Protocols Volume 1: Optical-Based Detectors, Methods in Molecular Biology; Rasooly, A., Prickril, B., Eds.; Springer Science + Business Media: Berlin, Germany, 2017; Volume 1571, pp. 251–266. [Google Scholar]


| PCR Assay | Genome Detection of | Primer/Probe | Sequence 5′–3′ | Amplicon (Base Pair) | Reference |
| Polci-mix | PPRV | PPR-Np-F298 | CGC CTT GTT GAG GTA GTT CAA AGT | 69 | Polci et al., 2015 [40] |
| PPR-Np-R366 | ATC AGC ACC ACG TGA TGC A | ||||
| PPR-NP-FAM-MGB | FAM– CAG TCC GGG TTG ACC T –MGBNFQ | ||||
| PPRV-mix 6 | PPRV | PPR-H-8502-F | GAC CTC CYT CAT TTT GCA ATG G | 85 | in this study |
| PPR-H-8586-R | ACT GAC YCT GAT CAC YCC GTA | ||||
| PPR-H-8538-FAM | FAM-CCC RTG GTC AGA RGG GAG AAT CCC-BHQ1 | ||||
| Righter | Mccp | Mccp-1F | CGC TCA CAT AGC CAA TCA TC | 152 | Righter et al., 2011 [43] |
| Mccp-1R | TCG TTT TTA AGA GAA AAT CAA GCA | ||||
| Mccp-1FAM | FAM-CAA GCT GAT GAA CAT AAA AAT GAT G-BHQ1 | ||||
| IRES-3C | FMDV | FMD-IRES-3.1F | CTG GWG RCA GGC TAA GGA T | 69 | Wernike et al., 2013 [44] modified |
| FMD-IRES-3R | CCC TTC TCA GAT YCC RAG TG | ||||
| FMD-IRES-3FAM | FAM-CCC TTC AGG TAC CCC GAG GTA ACA-BHQ1 | ||||
| Parapox-B2L | Parapoxvirus | PPV-B2L-455F | TCG ATG CGG TGC AGC AC | 95 | Nitsche et al., 2006 [45] |
| PPV-B2L-539R | GCG GCG TAT TCT TCT CGG AC | ||||
| PPV-B2L-FAM-MGB | FAM-TGC GGT AGA AGC C-MGB | ||||
| Capri-p32-mix1 | Capripoxvirus | Capri-p32for | AAA ACG GTA TAT GGA ATA GAG TTG GAA | 89 | Bowden et al., 2008 [46] modified; Dietze et al. 2018 [47] |
| Capri-p32rev | AAA TGA AAC CAA TGG ATG GGA TA | ||||
| Capri-p32-FAM | FAM-ATG GAT GGC TCA TAG ATT TCC TGA T-BHQ1 | ||||
| EGFP-mix 1 | Enhanced green fluorescent protein gene | EGFP-1-F | GAC CAC TAC CAG CAG AAC AC | 132 | Hoffmann et al., 2006 [48] |
| EGFP-2-R | GAA CTC CAG CAG GAC CAT G | ||||
| EGFP-FAM | FAM-AGC ACC CAG TCC GCC CTG AGC A-BHQ1 | ||||
| β-Actin-DNA-mix 2 | beta-actin mRNA | ACT-1030-F | AGC GCA AGT ACT CCG TGT G | 106 | Toussaint et al., 2007 [49] modified; Wernike et al., 2011 [50] |
| ACT-1135-R | CGG ACT CAT CGT ACT CCT GCT T | ||||
| ACT-1081-FAM | FAM-TCG CTG TCC ACC TTC CAG CAG ATG T-BHQ1 |
| CFX96 Touch | AriaMx | MIC | LightCycler 2.0 | LightCycler 96 | Liberty16 | |
|---|---|---|---|---|---|---|
| Simple Handling of the Software | ||||||
| Intuitive | +++ | ++ | ++ | - | + | ++ |
| On-board instrument diagnostics 1 | Yes | Yes | No | No | Yes | No |
| Touch-screen option | Yes | Yes | No | No | Yes | Yes |
| Cycler Equipment | ||||||
| Samples per instrument | 96 | 96 | 48 | 32 | 96 | 16 |
| Reaction vessels used | 96-well plate | 96-well plate | 4 tube stripes | glass capillaries | 96-well plate | 8 strip PCR tube |
| Ramping rates (°C/s) | 3.3–5.0 | 2.5–6.0 | 4.0–5.0 | 0.1–20 | 2.2–4.4 | 2.3 |
| Heating (°C/s) | n.s. | 6.0 | 5.0 | n.s. | 4.4 | n.s. |
| Cooling (°C/s) | n.s. | 2.5–3.0 | 4.0 | n.s. | 2.2 | n.s. |
| Power supply | External | external | External | External | external | external and battery |
| Dimensions | ||||||
| Width × deep × height (cm) | 33 × 46 × 36 | 50 × 46 × 42 | 15 × 15 × 13 | 28 × 39 × 51 | 40 × 40 × 53 | 11 × 21 × 12 |
| Weight (kg) | 21 | 23 | 2.1 | 22 | 27 | 3.2 |
| Duration of a Single Run | ||||||
| Standard protocol | 1 h 38 min | 1 h 32 min | 1 h 38 min | 1 h 23 min | 1 h 34 min | 1 h 40 min |
| Short protocol 5 | 38 min | 33 min | 39 min | 25 min | 34 min | 41 min |
| Takyon Kit | Qscript Kit | Capital Kit | |
|---|---|---|---|
| Storage Conditions | |||
| Storage at … temperature | 15–35 °C | room ~ | room ~ |
| Stability at room temperature for/until | 18 months | 9 months | expiry date |
| Storage after dissolution at … temperature | 4 °C (for 24 h) | n.s. | −20 °C |
| Features of the Kit (Manufacturer Specifications) | |||
| Reactions per kit (smallest size) | 50 | 8 | 200 |
| Recommended reaction size | 20 µL | 25 µL | 20 µL |
| Smallest number of samples after dissolution | 50 | 1 | 50 |
| Delivery format of the lyophilizate | one tube | 8-strip tubes | one tube |
| (A) Extraction: Original Protocol (17 min); RT-qPCR: Standard Protocol on BioRad CFX96 (1 h 38 min) | ||||||||
| Pathogen Detection | Control Assays | |||||||
| PPRV | FMDV | Parapoxvirus | Capripoxvirus | Mccp | EGFP-1-FAM | β-Actin-DNA-2-FAM | Non-Oligo control | |
| Single Infection | ||||||||
| Ivory Coast/89 (LI) | 20.3 | No Cq | No Cq | No Cq | No Cq | 28.1 | 31.7 | No Cq |
| Nigeria 75/1 (LII) | 23.8 | No Cq | No Cq | No Cq | No Cq | 27.7 | 34 | No Cq |
| Sudan/72 (LIII) | 24.1 | No Cq | No Cq | No Cq | No Cq | 27.4 | 34.9 | No Cq |
| Kurdistan/2011 (LIV) | 22.1 | No Cq | No Cq | No Cq | No Cq | 27.3 | 36 | No Cq |
| Indien/Shahjadpur (LIV) | 19.3 | No Cq | No Cq | No Cq | No Cq | 27.8 | 34.1 | No Cq |
| SMRV/UAE/2018/V135/Dubai (LIV) | 25.2 | No Cq | No Cq | No Cq | No Cq | 27.5 | 34.5 | No Cq |
| FMDV (A Iran 8/2015) | No Cq | 24.8 | No Cq | No Cq | No Cq | 26.7 | 36.1 | No Cq |
| Parapoxvirus ovis | No Cq | No Cq | 29 | No Cq | No Cq | 30.8 | 33.9 | No Cq |
| GTPV (Indian) | No Cq | No Cq | No Cq | 28 | No Cq | 26.3 | 37.2 | No Cq |
| Mccp | No Cq | No Cq | No Cq | No Cq | 20 | 26.5 | 28.3 | No Cq |
| Mixed Infection | ||||||||
| PPRV * + Mccp | 23 | No Cq | No Cq | No Cq | 31.1 | 27.1 | 32.4 | No Cq |
| PPRV * + FMDV | 24.9 | 29.1 | No Cq | No Cq | No Cq | 27.5 | 35.1 | No Cq |
| PPRV * + GTPV | 30.1 | No Cq | No Cq | 24.5 | No Cq | 26.8 | 34.2 | No Cq |
| FMDV + Mccp | No Cq | 29.6 | No Cq | No Cq | 23.2 | 27.5 | 31.2 | No Cq |
| GTPV + Parapoxvirus ovis | No Cq | No Cq | 31.5 | 25.3 | No Cq | 26.8 | 34.2 | No Cq |
| PPRV * + Mccp + Parapoxvirus ovis | 30.9 | No Cq | 30.5 | No Cq | 22.4 | 26.3 | 30.1 | No Cq |
| (B) Extraction: Short Protocol (7 min); RT-qPCR: Standard Protocol on BioRad CFX96 (1 h 38 min) | ||||||||
| Pathogen Detection | Control Assays | |||||||
| PPRV | FMDV | Parapoxvirus | Capripoxvirus | Mccp | EGFP-1-FAM | β-Actin-DNA-2-FAM | Non-Oligo control | |
| Single Infection | ||||||||
| Ivory Coast/89 (LI) | 21 | No Cq | No Cq | No Cq | No Cq | 25.5 | 30.5 | No Cq |
| Nigeria 75/1 (LII) | 24.4 | No Cq | No Cq | No Cq | No Cq | 26 | 34.8 | No Cq |
| Sudan/72 (LIII) | 24.5 | No Cq | No Cq | No Cq | No Cq | 25.7 | 34 | No Cq |
| Kurdistan/2011 (LIV) | 23 | No Cq | No Cq | No Cq | No Cq | 25.4 | 32.7 | No Cq |
| Indien/Shahjadpur (LIV) | 19.5 | No Cq | No Cq | No Cq | No Cq | 25.4 | 31.4 | No Cq |
| SMRV/UAE/2018/V135/Dubai (LIV) | 25.6 | No Cq | No Cq | No Cq | No Cq | 25.4 | 33.4 | No Cq |
| (C) Extraction: Original Protocol (17 min); RT-qPCR: Short Protocol on BioRad CFX96 (35 min) | ||||||||
| Single Infection | ||||||||
| Ivory Coast/89 (LI) | 21.8 | No Cq | No Cq | No Cq | No Cq | 26.3 | 31 | No Cq |
| Nigeria 75/1 (LII) | 26.1 | No Cq | No Cq | No Cq | No Cq | 25.3 | 34.3 | No Cq |
| Sudan/72 (LIII) | 27.1 | No Cq | No Cq | No Cq | No Cq | 25.2 | 35.6 | No Cq |
| Kurdistan/2011 (LIV) | 24.6 | No Cq | No Cq | No Cq | No Cq | 25.2 | 35.1 | No Cq |
| Indien/Shahjadpur (LIV) | 21.5 | No Cq | No Cq | No Cq | No Cq | 25.5 | 33 | No Cq |
| SMRV/UAE/2018/V135/Dubai (LIV) | 27.1 | No Cq | No Cq | No Cq | No Cq | 25.9 | 36.3 | No Cq |
| (D) Extraction: Short Protocol (7 min); RT-qPCR: Short Protocol on BioRad CFX96 (35 min) | ||||||||
| Pathogen Detection | Control Assays | |||||||
| PPRV | FMDV | Parapoxvirus | Capripoxvirus | Mccp | EGFP-1-FAM | β-Actin-DNA-2-FAM | Non-Oligo control | |
| Single Infection | ||||||||
| Ivory Coast/89 (LI) | 23.4 | No Cq | No Cq | No Cq | No Cq | 26.3 | 33 | No Cq |
| Nigeria 75/1 (LII) | 27.2 | No Cq | No Cq | No Cq | No Cq | 25.9 | 37.3 | No Cq |
| Sudan/72 (LIII) | 29 | No Cq | No Cq | No Cq | No Cq | 25.9 | 38.1 | No Cq |
| Kurdistan/2011 (LIV) | 26 | No Cq | No Cq | No Cq | No Cq | 25.9 | 36.2 | No Cq |
| Indien/Shahjadpur (LIV) | 22.8 | No Cq | No Cq | No Cq | No Cq | 25.8 | 36 | No Cq |
| SMRV/UAE/2018/V135/Dubai (LIV) | 28.1 | No Cq | No Cq | No Cq | No Cq | 26.1 | 38.5 | No Cq |
| FMDV (A Iran 8/2015) | No Cq | 27.5 | No Cq | No Cq | No Cq | 26.2 | 36.2 | No Cq |
| Parapoxvirus ovis | No Cq | No Cq | 31.4 | No Cq | No Cq | 26.4 | 35.6 | No Cq |
| GTPV (Indian) | No Cq | No Cq | No Cq | 29 | No Cq | 26.9 | 38.1 | No Cq |
| Mccp | No Cq | No Cq | No Cq | No Cq | 21.8 | 28.1 | 31.8 | No Cq |
| Mixed Infection | ||||||||
| PPRV * + Mccp | 26.3 | No Cq | No Cq | No Cq | 32.3 | 27.1 | 36.2 | No Cq |
| PPRV * + FMDV | 28.6 | 31.7 | No Cq | No Cq | No Cq | 27.2 | 38.2 | No Cq |
| PPRV * + GTPV | 34.7 | No Cq | No Cq | 26.5 | No Cq | 27.3 | 36.9 | No Cq |
| FMDV + Mccp | No Cq | 32.8 | No Cq | No Cq | 24.4 | 28.2 | 34.2 | No Cq |
| GTPV + Parapoxvirus ovis | No Cq | No Cq | 33.1 | 27.5 | No Cq | 27.1 | 37.4 | No Cq |
| PPRV * + Mccp + Parapoxvirus ovis | 34.5 | No Cq | 31.7 | No Cq | 24 | 27.6 | 33 | No Cq |
| (A) Extraction: Original Protocol (17 min); RT-qPCR: Standard Protocol on Liberty16 (1 h 40 min) | ||||||||
| Pathogen Detection | Control Assays | |||||||
| PPRV | FMDV | Parapoxvirus | Capripoxvirus | Mccp | EGFP-1-FAM | β-Actin-DNA-2-FAM | Non-Oligo control | |
| Single Infection | ||||||||
| Ivory Coast/89 (LI) | 22.6 | No Cq | No Cq | No Cq | No Cq | 29.4 | 30.3 | No Cq |
| Nigeria 75/1 (LII) | 26.1 | No Cq | No Cq | No Cq | No Cq | 29.1 | 34.6 | No Cq |
| Sudan/72 (LIII) | 25.5 | No Cq | No Cq | No Cq | No Cq | 28.8 | 34.5 | No Cq |
| Kurdistan/2011 (LIV) | 24.5 | No Cq | No Cq | No Cq | No Cq | 29.2 | 33.4 | No Cq |
| Indien/Shahjadpur (LIV) | 21.8 | No Cq | No Cq | No Cq | No Cq | 30.2 | 32.1 | No Cq |
| SMRV/UAE/2018/V135/Dubai (LIV) | 26.8 | No Cq | No Cq | No Cq | No Cq | 28.7 | 34.5 | No Cq |
| FMDV (A Iran 8/2015) | No Cq | 24.7 | No Cq | No Cq | No Cq | 29.2 | 34.1 | No Cq |
| Parapoxvirus ovis | No Cq | No Cq | 30.9 | No Cq | No Cq | 32.8 | 34.5 | No Cq |
| GTPV (Indian) | No Cq | No Cq | No Cq | 29.7 | No Cq | 28.5 | 36 | No Cq |
| Mccp | No Cq | No Cq | No Cq | No Cq | 22.8 | 29.3 | 29.2 | No Cq |
| Mixed Infection | ||||||||
| PPRV * + Mccp | 25.6 | No Cq | No Cq | No Cq | 32 | 28.6 | 34.4 | No Cq |
| PPRV * + FMDV | 26.5 | 29.8 | No Cq | No Cq | No Cq | 29.8 | 35.7 | No Cq |
| PPRV * + GTPV | 32.8 | No Cq | No Cq | 25.9 | No Cq | 28.8 | 34.5 | No Cq |
| FMDV + Mccp | No Cq | 29.2 | No Cq | No Cq | 21.9 | 29.6 | 30 | No Cq |
| GTPV + Parapoxvirus ovis | No Cq | No Cq | 32.5 | 26.9 | No Cq | 29 | 33.3 | No Cq |
| PPRV * + Mccp + Parapoxvirus ovis | 32.1 | No Cq | 32.2 | No Cq | 23.9 | 29.6 | 30.9 | No Cq |
| (B) Extraction: Short Protocol (7 min); RT-qPCR: Short Protocol on Liberty16 (41 min) | ||||||||
| Pathogen Detection | Control Assays | |||||||
| PPRV | FMDV | Parapoxvirus | Capripoxvirus | Mccp | EGFP-1-FAM | β-Actin-DNA-2-FAM | Non-Oligo control | |
| Single Infection | ||||||||
| Ivory Coast/89 (LI) | 25.8 | No Cq | No Cq | No Cq | No Cq | 29.7 | 34.8 | No Cq |
| Nigeria 75/1 (LII) | 30.3 | No Cq | No Cq | No Cq | No Cq | 29.4 | 35.2 | No Cq |
| Sudan/72 (LIII) | 30.4 | No Cq | No Cq | No Cq | No Cq | 27.9 | 36.5 | No Cq |
| Kurdistan/2011 (LIV) | 28.4 | No Cq | No Cq | No Cq | No Cq | 30 | 35.9 | No Cq |
| Indien/Shahjadpur (LIV) | 25.1 | No Cq | No Cq | No Cq | No Cq | 29.8 | 35.5 | No Cq |
| SMRV/UAE/2018/V135/Dubai (LIV) | 30.6 | No Cq | No Cq | No Cq | No Cq | 29.8 | No Cq | No Cq |
| FMDV (A Iran 8/2015) | No Cq | 26.9 | No Cq | No Cq | No Cq | 29.2 | 35.1 | No Cq |
| Parapoxvirus ovis | No Cq | No Cq | 32.6 | No Cq | No Cq | 32 | 34.9 | No Cq |
| GTPV (Indian) | No Cq | No Cq | No Cq | 31.1 | No Cq | 33.7 | No Cq | No Cq |
| Mccp | No Cq | No Cq | No Cq | No Cq | 22.5 | 33.5 | 32.4 | No Cq |
| Mixed Infection | ||||||||
| PPRV * + Mccp | 27.8 | No Cq | No Cq | No Cq | 32.2 | 29.4 | 36.7 | No Cq |
| PPRV * + FMDV | 29.2 | 30.9 | No Cq | No Cq | No Cq | 31.5 | 35.5 | No Cq |
| PPRV * + GTPV | 36 | No Cq | No Cq | 27.9 | No Cq | 31 | 35.7 | No Cq |
| FMDV + Mccp | No Cq | 31.8 | No Cq | No Cq | 25.5 | 29.9 | 33.7 | No Cq |
| GTPV + Parapoxvirus ovis | No Cq | No Cq | 33.6 | 28.8 | No Cq | 30.7 | 34.7 | No Cq |
| PPRV * + Mccp + Parapoxvirus ovis | 34.9 | No Cq | 32.5 | No Cq | 24.2 | 29.8 | 31.9 | No Cq |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halecker, S.; Mettenleiter, T.C.; Beer, M.; Hoffmann, B. “FastCheckFLI PPR-like”—A Molecular Tool for the Fast Genome Detection of PPRV and Differential Diagnostic Pathogens. Viruses 2020, 12, 1227. https://doi.org/10.3390/v12111227
Halecker S, Mettenleiter TC, Beer M, Hoffmann B. “FastCheckFLI PPR-like”—A Molecular Tool for the Fast Genome Detection of PPRV and Differential Diagnostic Pathogens. Viruses. 2020; 12(11):1227. https://doi.org/10.3390/v12111227
Chicago/Turabian StyleHalecker, Sabrina, Thomas C. Mettenleiter, Martin Beer, and Bernd Hoffmann. 2020. "“FastCheckFLI PPR-like”—A Molecular Tool for the Fast Genome Detection of PPRV and Differential Diagnostic Pathogens" Viruses 12, no. 11: 1227. https://doi.org/10.3390/v12111227
APA StyleHalecker, S., Mettenleiter, T. C., Beer, M., & Hoffmann, B. (2020). “FastCheckFLI PPR-like”—A Molecular Tool for the Fast Genome Detection of PPRV and Differential Diagnostic Pathogens. Viruses, 12(11), 1227. https://doi.org/10.3390/v12111227





