HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites
Abstract
:1. Introduction
2. Materials and Methods
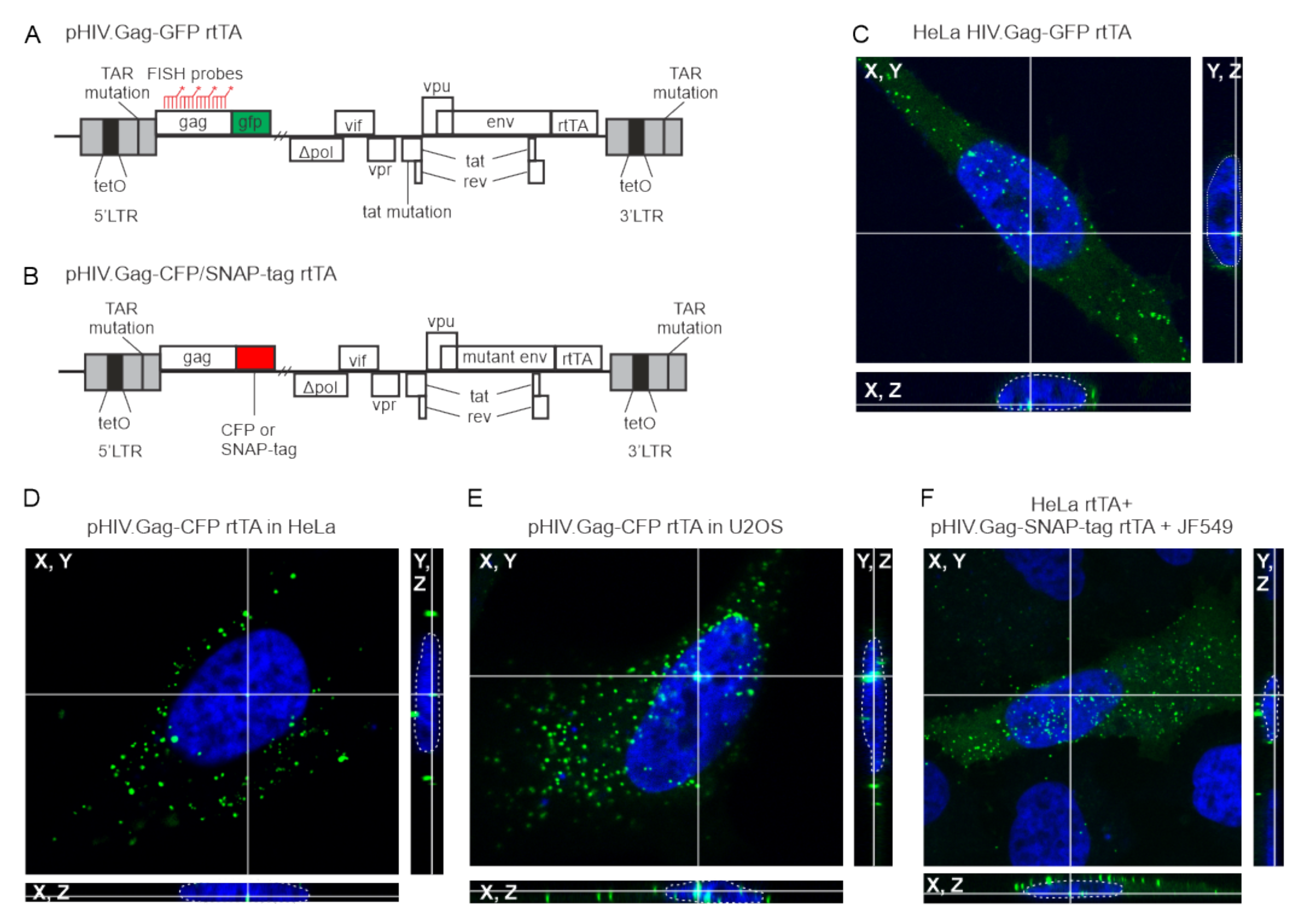
2.1. Plasmids, Cell Culture, and Transfections
2.2. Dox-Induction and Visualization of USvRNA and GAPDH RNA in HeLa and U2OS Cells
2.3. Prostratin Induction and Sequential IF/FISH Labeling of USvRNA and Gag in JLat 10.6 Cells
2.4. Confocal Microscopy
2.5. Quantitative Image Analysis
2.6. Subcellular Fractionations
2.7. RNA-Immunoprecipitation and Biochemical Analysis
3. Results
3.1. HIV-1 Gag Forms Nuclear Foci in Multiple Cell Types
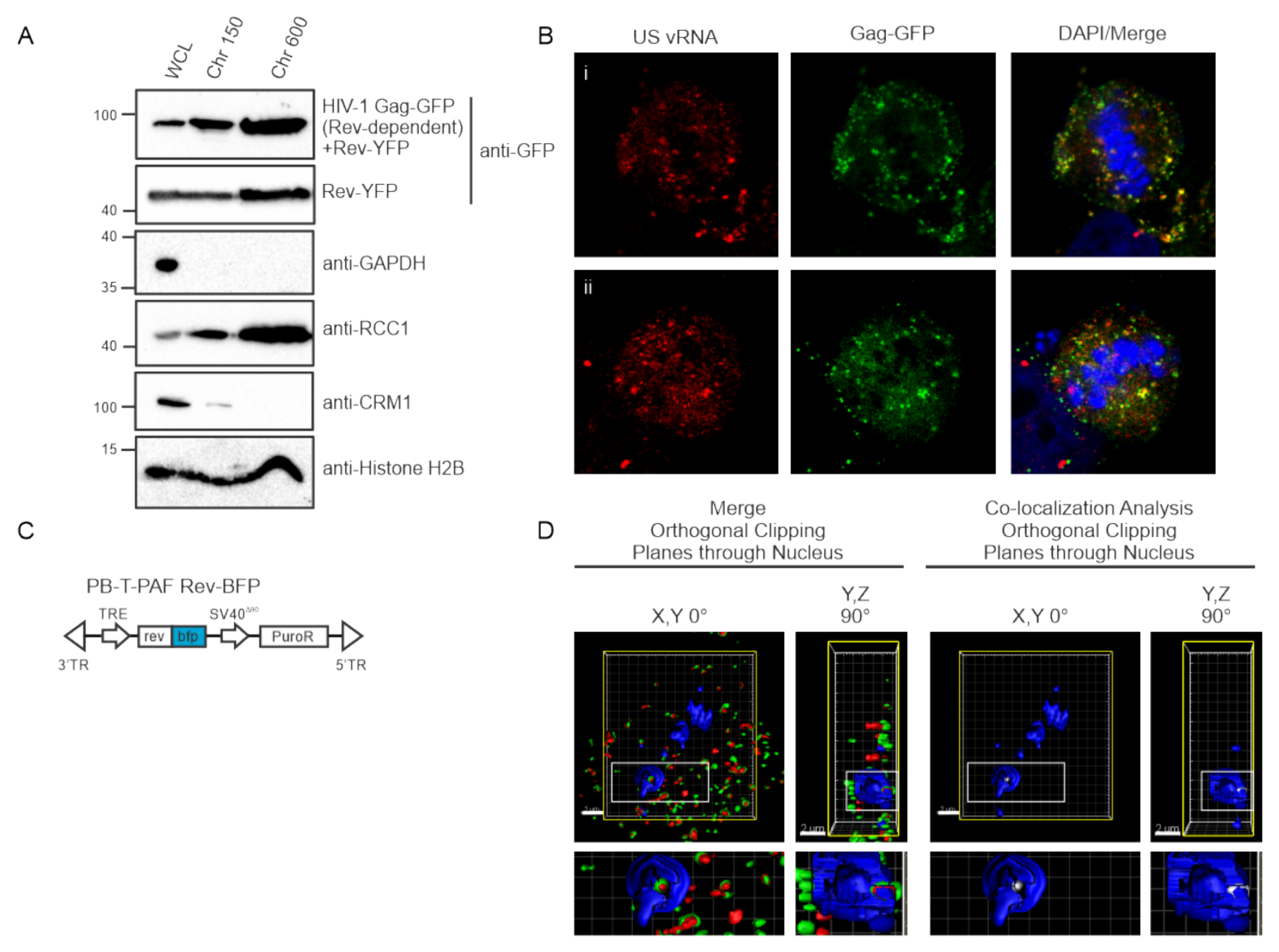
3.2. HIV-1 Gag Is Present within the Nuclear Fractions Using Biochemical Methods
3.3. HIV-1 Gag Specifically co-Localizes with USvRNA in Discrete Nuclear Foci
3.4. HIV-1 Gag Associates with USvRNA in the Nucleus
3.5. Effects of Transcription Inhibition on HIV-1 Gag/USvRNA Co-Localization
3.6. Effects of LMB Treatment on HIV-1 Gag/USvRNA Association
3.7. Nuclear and Chromatin Associated Localization of HIV-1 Gag and Rev
3.8. Three-Dimensional Co-Localization of HIV-1 Gag, USvRNA, and Rev
3.9. Co-Localization of HIV-1 Gag and USvRNA in JLat 10.6 CD4+ T Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rabson, A.B.; Graves, B.J. Synthesis and Processing of Viral RNA. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Laine, S.; Pessel-Vivares, L.; Mougel, M. Cell biology of retroviral RNA packaging. RNA Biol. 2011, 8, 572–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jouvenet, N.; Neil, S.J.; Bess, C.; Johnson, M.C.; Virgen, C.A.; Simon, S.M.; Bieniasz, P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006, 4, e435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barajas, B.C.; Tanaka, M.; Robinson, B.A.; Phuong, D.J.; Chutiraka, K.; Reed, J.C.; Lingappa, J.R. Identifying the assembly intermediate in which Gag first associates with unspliced HIV-1 RNA suggests a novel model for HIV-1 RNA packaging. PLoS Pathog. 2018, 14, e1006977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swanson, C.M.; Malim, M.H. Retrovirus RNA trafficking: From chromatin to invasive genomes. Traffic 2006, 7, 1440–1450. [Google Scholar] [CrossRef]
- Milev, M.P.; Brown, C.M.; Mouland, A.J. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology 2010, 7, 41. [Google Scholar] [CrossRef] [Green Version]
- Garbitt-Hirst, R.; Kenney, S.P.; Parent, L.J. Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J. Virol. 2009, 83, 6790–6797. [Google Scholar] [CrossRef] [Green Version]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [Green Version]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef] [Green Version]
- Butterfield-Gerson, K.L.; Scheifele, L.Z.; Ryan, E.P.; Hopper, A.K.; Parent, L.J. Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 2006, 80, 1798–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenney, S.P.; Lochmann, T.L.; Schmid, C.L.; Parent, L.J. Intermolecular interactions between retroviral Gag proteins in the nucleus. J. Virol. 2008, 82, 683–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheifele, L.Z.; Kenney, S.P.; Cairns, T.M.; Craven, R.C.; Parent, L.J. Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J. Virol. 2007, 81, 10718–10728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheifele, L.Z.; Ryan, E.P.; Parent, L.J. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 2005, 79, 8732–8741. [Google Scholar] [CrossRef] [Green Version]
- Maldonado, R.J.K.; Rice, B.; Chen, E.C.; Tuffy, K.M.; Chiari, E.F.; Fahrbach, K.M.; Hope, T.J.; Parent, L.J. Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parent, L.J. New insights into the nuclear localization of retroviral Gag proteins. Nucleus 2011, 2, 92–97. [Google Scholar] [CrossRef] [Green Version]
- Grewe, B.; Hoffmann, B.; Ohs, I.; Blissenbach, M.; Brandt, S.; Tippler, B.; Grunwald, T.; Uberla, K. Cytoplasmic utilization of human immunodeficiency virus type 1 genomic RNA is not dependent on a nuclear interaction with gag. J. Virol. 2012, 86, 2990–3002. [Google Scholar] [CrossRef] [Green Version]
- Lochmann, T.L.; Bann, D.V.; Ryan, E.P.; Beyer, A.R.; Mao, A.; Cochrane, A.; Parent, L.J. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 2013, 171, 304–318. [Google Scholar] [CrossRef] [Green Version]
- Kemler, I.; Saenz, D.; Poeschla, E. Feline immunodeficiency virus Gag is a nuclear shuttling protein. J. Virol. 2012, 86, 8402–8411. [Google Scholar] [CrossRef] [Green Version]
- Baluyot, M.F.; Grosse, S.A.; Lyddon, T.D.; Janaka, S.K.; Johnson, M.C. CRM1-dependent trafficking of retroviral Gag proteins revisited. J. Virol. 2012, 86, 4696–4700. [Google Scholar] [CrossRef] [Green Version]
- Bohl, C.R.; Brown, S.M.; Weldon, R.A., Jr. The pp24 phosphoprotein of Mason-Pfizer monkey virus contributes to viral genome packaging. Retrovirology 2005, 2, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, A.R.; Bann, D.V.; Rice, B.; Pultz, I.S.; Kane, M.; Goff, S.P.; Golovkina, T.V.; Parent, L.J. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J. Virol. 2013, 87, 1069–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullers, E.; Stirnnagel, K.; Kaulfuss, S.; Lindemann, D. Prototype foamy virus gag nuclear localization: A novel pathway among retroviruses. J. Virol. 2011, 85, 9276–9285. [Google Scholar] [CrossRef] [Green Version]
- Wong, R.; Balachandran, A.; Mao, A.Y.; Dobson, W.; Gray-Owen, S.; Cochrane, A. Differential effect of CLK SR Kinases on HIV-1 gene expression: Potential novel targets for therapy. Retrovirology 2011, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Vink, M.; Berkhout, B.; Das, A.T. Modification of the Tet-On regulatory system prevents the conditional-live HIV-1 variant from losing doxycycline-control. Retrovirology 2006, 3, 82. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Vink, M.; Klaver, B.; Verhoef, K.; Marzio, G.; Das, A.T.; Berkhout, B. The genetic stability of a conditional live HIV-1 variant can be improved by mutations in the Tet-On regulatory system that restrain evolution. J. Biol. Chem. 2006, 281, 17084–17091. [Google Scholar] [CrossRef] [Green Version]
- Marzio, G.; Verhoef, K.; Vink, M.; Berkhout, B. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. USA 2001, 98, 6342–6347. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M.H.; Coates, C.J.; George, A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007, 15, 139–145. [Google Scholar] [CrossRef]
- Li, Z.; Michael, I.P.; Zhou, D.; Nagy, A.; Rini, J.M. Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. Proc. Natl. Acad. Sci. USA 2013, 110, 5004–5009. [Google Scholar] [CrossRef] [Green Version]
- Craven, R.C.; Leure-duPree, A.E.; Weldon, R.A., Jr.; Wills, J.W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 1995, 69, 4213–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pocock, G.M.; Becker, J.T.; Swanson, C.M.; Ahlquist, P.; Sherer, N.M. HIV-1 and M-PMV RNA Nuclear Export Elements Program Viral Genomes for Distinct Cytoplasmic Trafficking Behaviors. PLoS Pathog. 2016, 12, e1005565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermida-Matsumoto, L.; Resh, M.D. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 2000, 74, 8670–8679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemler, I.; Meehan, A.; Poeschla, E.M. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 2010, 84, 6352–6366. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Oda, K.; Yokota, S.; Takatsuki, A.; Ikehara, Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 1988, 263, 18545–18552. [Google Scholar] [PubMed]
- Jordan, A.; Bisgrove, D.; Verdin, E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003, 22, 1868–1877. [Google Scholar] [CrossRef] [Green Version]
- Matic, I.; van Hagen, M.; Schimmel, J.; Macek, B.; Ogg, S.C.; Tatham, M.H.; Hay, R.T.; Lamond, A.I.; Mann, M.; Vertegaal, A.C.O. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteom. 2008, 7, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Grimm, J.B.; English, B.P.; Chen, J.; Slaughter, J.P.; Zhang, Z.; Revyakin, A.; Patel, R.; Macklin, J.J.; Normanno, D.; Singer, R.H.; et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 2015, 12, 244–250. [Google Scholar] [CrossRef]
- Tsang, M.; Gantchev, J.; Ghazawi, F.M.; Litvinov, I.V. Protocol for adhesion and immunostaining of lymphocytes and other non-adherent cells in culture. Biotechniques 2017, 63, 230–233. [Google Scholar] [CrossRef] [Green Version]
- Raj, A.; van den Bogaard, P.; Rifkin, S.A.; van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef] [Green Version]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mensa, L.; Crespo, G.; Gastinger, M.J.; Kabat, J.; Perez-del-Pulgar, S.; Miquel, R.; Emerson, S.U.; Purcell, R.H.; Forns, X. Hepatitis C virus receptors claudin-1 and occludin after liver transplantation and influence on early viral kinetics. Hepatology 2011, 53, 1436–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, E.; Matthias, P.; Muller, M.M.; Schaffner, W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989, 17, 6419. [Google Scholar] [CrossRef] [Green Version]
- Henikoff, S.; Henikoff, J.G.; Sakai, A.; Loeb, G.B.; Ahmad, K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009, 19, 460–469. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nawroth, I.; Mueller, F.; Basyuk, E.; Beerens, N.; Rahbek, U.L.; Darzacq, X.; Bertrand, E.; Kjems, J.; Schmidt, U. Stable assembly of HIV-1 export complexes occurs cotranscriptionally. RNA 2014, 20, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacampo, S.; Cochrane, A. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 1996, 70, 8332–8339. [Google Scholar] [CrossRef] [Green Version]
- Niedojadlo, J.; Perret-Vivancos, C.; Kalland, K.H.; Cmarko, D.; Cremer, T.; van Driel, R.; Fakan, S. Transcribed DNA is preferentially located in the perichromatin region of mammalian cell nuclei. Exp. Cell. Res. 2011, 317, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Chakalova, L.; Fraser, P. Organization of transcription. Cold Spring Harb. Perspect Biol. 2010, 2, a000729. [Google Scholar] [CrossRef] [Green Version]
- Tobaly-Tapiero, J.; Bittoun, P.; Lehmann-Che, J.; Delelis, O.; Giron, M.L.; de The, H.; Saib, A. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 2008, 9, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Lesbats, P.; Serrao, E.; Maskell, D.P.; Pye, V.E.; O’Reilly, N.; Lindemann, D.; Engelman, A.N.; Cherepanov, P. Structural basis for spumavirus GAG tethering to chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, 5509–5514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elis, E.; Ehrlich, M.; Prizan-Ravid, A.; Laham-Karam, N.; Bacharach, E. p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog. 2012, 8, e1003103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wight, D.J.; Boucherit, V.C.; Nader, M.; Allen, D.J.; Taylor, I.A.; Bishop, K.N. The gammaretroviral p12 protein has multiple domains that function during the early stages of replication. Retrovirology 2012, 9, 83. [Google Scholar] [CrossRef] [Green Version]
- Schneider, W.M.; Brzezinski, J.D.; Aiyer, S.; Malani, N.; Gyuricza, M.; Bushman, F.D.; Roth, M.J. Viral DNA tethering domains complement replication-defective mutations in the p12 protein of MuLV Gag. Proc. Natl. Acad. Sci. USA 2013, 110, 9487–9492. [Google Scholar] [CrossRef] [Green Version]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef] [Green Version]
- Symons, J.; Chopra, A.; Malatinkova, E.; De Spiegelaere, W.; Leary, S.; Cooper, D.; Abana, C.O.; Rhodes, A.; Rezaei, S.D.; Vandekerckhove, L.; et al. HIV integration sites in latently infected cell lines: Evidence of ongoing replication. Retrovirology 2017, 14, 2. [Google Scholar] [CrossRef] [Green Version]
- Ukah, O.B.; Puray-Chavez, M.; Tedbury, P.R.; Herschhorn, A.; Sodroski, J.G.; Sarafianos, S.G. Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells. Viruses 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Kulkosky, J.; Culnan, D.M.; Roman, J.; Dornadula, G.; Schnell, M.; Boyd, M.R.; Pomerantz, R.J. Prostratin: Activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 2001, 98, 3006–3015. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.A.; Chen, L.F.; Kwon, H.; Fenard, D.; Bisgrove, D.; Verdin, E.; Greene, W.C. Prostratin antagonizes HIV latency by activating NF-kappaB. J. Biol. Chem. 2004, 279, 42008–42017. [Google Scholar] [CrossRef] [Green Version]
- Kulkosky, J.; Sullivan, J.; Xu, Y.; Souder, E.; Hamer, D.H.; Pomerantz, R.J. Expression of latent HAART-persistent HIV type 1 induced by novel cellular activating agents. AIDS Res. Hum. Retrovir. 2004, 20, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Puray-Chavez, M.; Tedbury, P.R.; Huber, A.D.; Ukah, O.B.; Yapo, V.; Liu, D.; Ji, J.; Wolf, J.J.; Engelman, A.N.; Sarafianos, S.G. Multiplex single-cell visualization of nucleic acids and protein during HIV infection. Nat. Commun. 2017, 8, 1882. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.E.; Malim, M.H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994, 8, 1538–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engeland, C.E.; Oberwinkler, H.; Schumann, M.; Krause, E.; Muller, G.A.; Krausslich, H.G. The cellular protein lyric interacts with HIV-1 Gag. J. Virol. 2011, 85, 13322–13332. [Google Scholar] [CrossRef] [Green Version]
- Engeland, C.E.; Brown, N.P.; Borner, K.; Schumann, M.; Krause, E.; Kaderali, L.; Muller, G.A.; Krausslich, H.G. Proteome analysis of the HIV-1 Gag interactome. Virology 2014, 460–461, 194–206. [Google Scholar] [CrossRef] [Green Version]
- Rye-McCurdy, T.; Olson, E.D.; Liu, S.; Binkley, C.; Reyes, J.P.; Thompson, B.R.; Flanagan, J.M.; Parent, L.J.; Musier-Forsyth, K. Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag. Viruses 2016, 8. [Google Scholar] [CrossRef]
- Le Sage, V.; Cinti, A.; Valiente-Echeverria, F.; Mouland, A.J. Proteomic analysis of HIV-1 Gag interacting partners using proximity-dependent biotinylation. Virol. J. 2015, 12, 138. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, C.; Cylinder, I.; Platt, E.J.; Barklis, E. Analysis of HIV-1 Gag protein interactions via biotin ligase tagging. J. Virol. 2015, 89, 3988–4001. [Google Scholar] [CrossRef] [Green Version]
- Blissenbach, M.; Grewe, B.; Hoffmann, B.; Brandt, S.; Uberla, K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J. Virol. 2010, 84, 6598–6604. [Google Scholar] [CrossRef] [Green Version]
- Arizala, J.; Rossi, J. Role of the Nucleolus in HIV Infection and Therapy. In The Nucleolus; Springer: New York, NY, USA, 2011; Volume 15, pp. 381–402. [Google Scholar]
- Michienzi, A.; Cagnon, L.; Bahner, I.; Rossi, J.J. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. USA 2000, 97, 8955–8960. [Google Scholar] [CrossRef] [Green Version]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Shinkai, H. Leptomycin B reduces matrix metalloproteinase-9 expression and suppresses cutaneous inflammation. J. Investig. Dermatol. 2005, 124, 331–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchive, C.; Roudier, F.; Castaings, L.; Brehaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.C.; Munoz-Najar, U.; Paik, J.H.; Claffey, K.; Yoshida, M.; Hla, T. Leptomycin B, an inhibitor of the nuclear export receptor CRM1, inhibits COX-2 expression. J. Biol. Chem. 2003, 278, 2773–2776. [Google Scholar] [CrossRef] [Green Version]
- Grewe, B.; Ehrhardt, K.; Hoffmann, B.; Blissenbach, M.; Brandt, S.; Uberla, K. The HIV-1 Rev protein enhances encapsidation of unspliced and spliced, RRE-containing lentiviral vector RNA. PLoS ONE 2012, 7, e48688. [Google Scholar] [CrossRef]
- Taniguchi, I.; Mabuchi, N.; Ohno, M. HIV-1 Rev protein specifies the viral RNA export pathway by suppressing TAP/NXF1 recruitment. Nucleic Acids Res. 2014, 42, 6645–6658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochrane, A. How does the journey affect the message(RNA)? RNA Biol. 2009, 6, 169–170. [Google Scholar] [CrossRef] [Green Version]
- Carmody, S.R.; Wente, S.R. mRNA nuclear export at a glance. J. Cell. Sci. 2009, 122, 1933–1937. [Google Scholar] [CrossRef] [Green Version]
- Butsch, M.; Boris-Lawrie, K. Destiny of unspliced retroviral RNA: Ribosome and/or virion? J. Virol. 2002, 76, 3089–3094. [Google Scholar] [CrossRef] [Green Version]
- Forget, A.; Chartrand, P. Cotranscriptional assembly of mRNP complexes that determine the cytoplasmic fate of mRNA. Transcription 2011, 2, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Knuckles, P.; Carl, S.H.; Musheev, M.; Niehrs, C.; Wenger, A.; Buhler, M. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 2017, 24, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.; Wu, B.; Nikolaitchik, O.A.; Mohan, P.R.; Pathak, V.K.; Hu, W.S. Visualizing the translation and packaging of HIV-1 full-length RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 6145–6155. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses 2020, 12, 1281. https://doi.org/10.3390/v12111281
Tuffy KM, Maldonado RJK, Chang J, Rosenfeld P, Cochrane A, Parent LJ. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses. 2020; 12(11):1281. https://doi.org/10.3390/v12111281
Chicago/Turabian StyleTuffy, Kevin M., Rebecca J. Kaddis Maldonado, Jordan Chang, Paul Rosenfeld, Alan Cochrane, and Leslie J. Parent. 2020. "HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites" Viruses 12, no. 11: 1281. https://doi.org/10.3390/v12111281
APA StyleTuffy, K. M., Maldonado, R. J. K., Chang, J., Rosenfeld, P., Cochrane, A., & Parent, L. J. (2020). HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses, 12(11), 1281. https://doi.org/10.3390/v12111281





