High Prevalence of Rotavirus A in Raw Sewage Samples from Northeast Spain
Abstract
:1. Introduction
2. Material and Methods
2.1. Raw Sewage Samples
2.2. Animal Fecal Samples
2.3. Virus Concentration
2.4. Nucleic Acid Extraction, Reverse Transcription (RT) and Quantitative PCR (qPCR)
2.5. VP4, VP6 and VP7 Amplification
2.6. Sequencing of the VP6 RT-PCR Products
2.7. Statistical Analysis
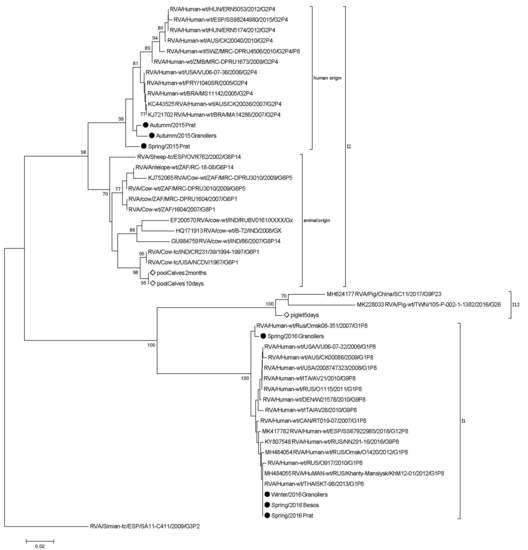
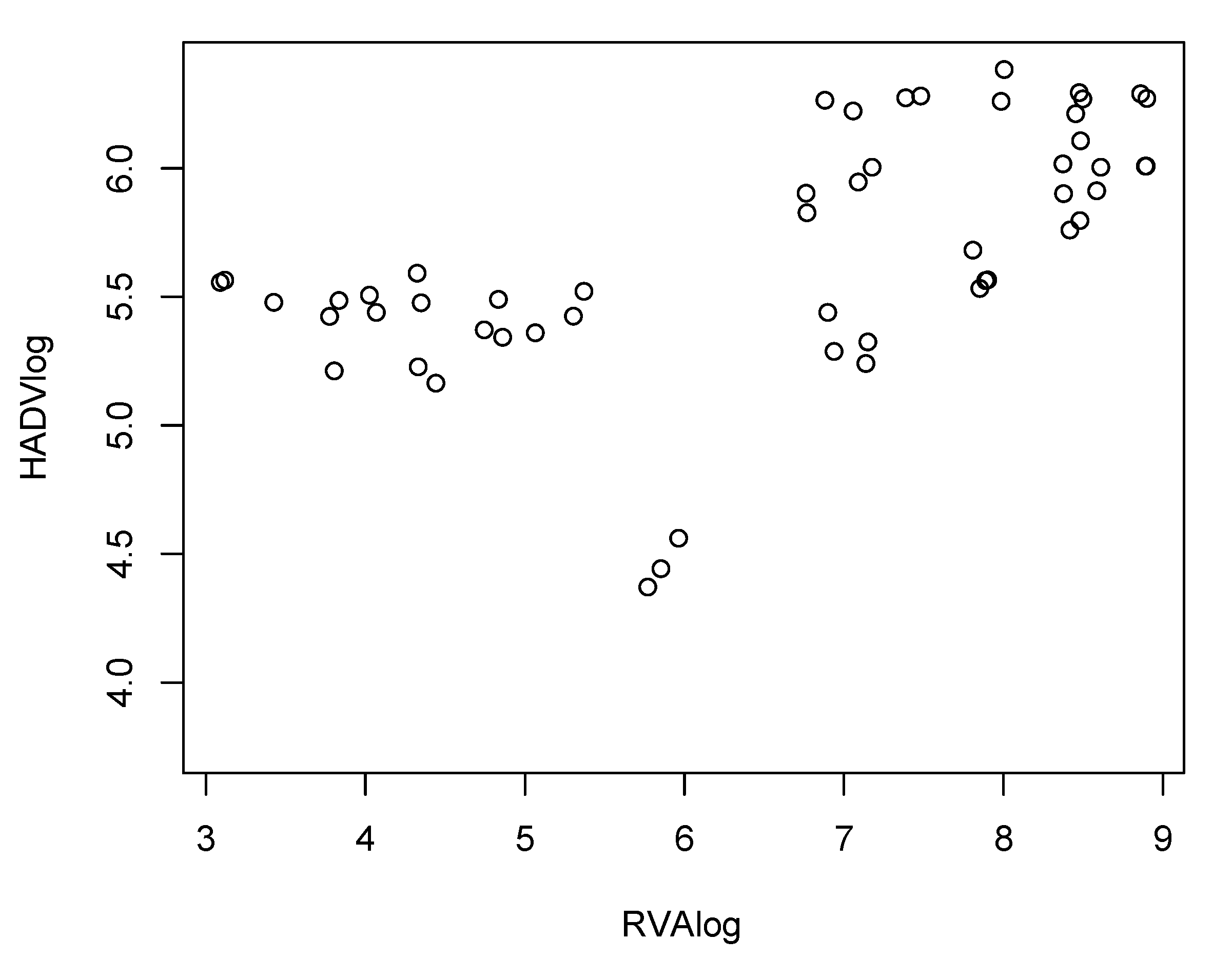
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization-Coordinated Global Rotavirus Surveillance Network. Global, Regional, and National Estimates of Rotavirus Mortality in Children. Clin. Infect. Dis. 2016, 62 (Suppl. S2), S96–S105. [Google Scholar] [CrossRef] [Green Version]
- Diez-Domingo, J.; Garces-Sanchez, M.; Gimenez-Sanchez, F.; Colomina-Rodriguez, J.; Martinon-Torres, F. [What have we learnt about rotavirus in Spain in the last 10 years?]. An. Pediatr. (Barc.) 2019. [Google Scholar] [CrossRef]
- Rahman, M.; Matthijnssens, J.; Yang, X.; Delbeke, T.; Arijs, I.; Taniguchi, K.; Iturriza-Gomara, M.; Iftekharuddin, N.; Azim, T.; Van Ranst, M. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol. 2007, 81, 2382–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthijnssens, J.; Van Ranst, M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr. Opin. Virol. 2012, 2, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Trojnar, E.; Sachsenroder, J.; Twardziok, S.; Reetz, J.; Otto, P.H.; Johne, R. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J. Gen. Virol. 2013, 94, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Bucardo, F.; Rippinger, C.M.; Svensson, L.; Patton, J.T. Vaccine-derived NSP2 segment in rotaviruses from vaccinated children with gastroenteritis in Nicaragua. Infect. Genet. Evol. 2012, 12, 1282–1294. [Google Scholar] [CrossRef] [Green Version]
- Delogu, R.; Ianiro, G.; Camilloni, B.; Fiore, L.; Ruggeri, F.M. Unexpected spreading of G12P[8] rotavirus strains among young children in a small area of central Italy. J. Med. Virol. 2015, 87, 1292–1302. [Google Scholar] [CrossRef]
- da Silva, M.F.; Fumian, T.M.; de Assis, R.M.; Fialho, A.M.; Carvalho-Costa, F.A.; da Silva Ribeiro de Andrade, J.; Leite, J.P. VP7 and VP8* genetic characterization of group A rotavirus genotype G12P[8]: Emergence and spreading in the Eastern Brazilian coast in 2014. J. Med. Virol. 2017, 89, 64–70. [Google Scholar] [CrossRef]
- Martella, V.; Banyai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef] [Green Version]
- Carter, M.J. Enterically infecting viruses: Pathogenicity, transmission and significance for food and waterborne infection. J. Appl. Microbiol. 2005, 98, 1354–1380. [Google Scholar] [CrossRef]
- Bosch, A.; Guix, S.; Sano, D.; Pinto, R.M. New tools for the study and direct surveillance of viral pathogens in water. Curr. Opin. Biotechnol. 2008, 19, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Heaton, P.M.; Ciarlet, M. Vaccines: The pentavalent rotavirus vaccine: Discovery to licensure and beyond. Clin. Infect. Dis. 2007, 45, 1618–1624. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Perez, D.; Alvarez Garcia, F.J.; Alvarez Aldean, J.; Cilleruelo Ortega, M.J.; Garces Sanchez, M.; Garcia Sanchez, N.; Hernandez Merino, A.; Mendez Hernandez, M.; Merino Moina, M.; Montesdeoca Melian, A.; et al. En representación del Comité Asesor de Vacunas de la Asociación Española de Pediatría (CAV-AEP), Immunisation schedule of the Spanish Association of Paediatrics: 2018 recommendations. An. Pediatr. (Barc.) 2018, 88, 53. [Google Scholar] [CrossRef] [PubMed]
- Pina, S.; Jofre, J.; Emerson, S.U.; Purcell, R.H.; Girones, R. Characterization of a strain of infectious hepatitis E virus isolated from sewage in an area where hepatitis E is not endemic. Appl. Environ. Microbiol. 1998, 64, 4485–4488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calgua, B.; Carratala, A.; Guerrero-Latorre, L.; de Abreu Correa, A.; Kohn, T.; Sommer, R.; Girones, R. UVC Inactivation of dsDNA and ssRNA Viruses in Water: UV Fluences and a qPCR-Based Approach to Evaluate Decay on Viral Infectivity. Food Environ. Virol. 2014, 6, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.Q.; Halkosalo, A.; Salminen, M.; Szakal, E.D.; Puustinen, L.; Vesikari, T. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J. Virol. Methods 2008, 153, 238–240. [Google Scholar] [CrossRef]
- Bofill-Mas, S.; Albinana-Gimenez, N.; Clemente-Casares, P.; Hundesa, A.; Rodriguez-Manzano, J.; Allard, A.; Calvo, M.; Girones, R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006, 72, 7894–7896. [Google Scholar] [CrossRef] [Green Version]
- Iturriza Gomara, M.; Wong, C.; Blome, S.; Desselberger, U.; Gray, J. Molecular characterization of VP6 genes of human rotavirus isolates: Correlation of genogroups with subgroups and evidence of independent segregation. J. Virol. 2002, 76, 6596–6601. [Google Scholar] [CrossRef] [Green Version]
- Gouvea, V.; Glass, R.I.; Woods, P.; Taniguchi, K.; Clark, H.F.; Forrester, B.; Fang, Z.Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 1990, 28, 276–282. [Google Scholar] [CrossRef] [Green Version]
- Gentsch, J.R.; Glass, R.I.; Woods, P.; Gouvea, V.; Gorziglia, M.; Flores, J.; Das, B.K.; Bhan, M.K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1365–1373. [Google Scholar] [CrossRef] [Green Version]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999; pp. 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Computing 2016. [Google Scholar] [CrossRef]
- Pohlert, T. The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R Package 2014, 27, 9. [Google Scholar]
- Fumian, T.M.; Leite, J.P.; Rose, T.L.; Prado, T.; Miagostovich, M.P. One year environmental surveillance of rotavirus specie A (RVA) genotypes in circulation after the introduction of the Rotarix(R) vaccine in Rio de Janeiro, Brazil. Water Res. 2011, 45, 5755–5763. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.M.; Bonomo, P.; Ianiro, G.; Battistone, A.; Delogu, R.; Germinario, C.; Chironna, M.; Triassi, M.; Campagnuolo, R.; Cicala, A.; et al. Rotavirus genotypes in sewage treatment plants and in children hospitalized with acute diarrhea in Italy in 2010 and 2011. Appl. Environ. Microbiol. 2015, 81, 241–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, N.; Lv, D.; Wang, S.; Lin, X.; Bi, Z.; Wang, H.; Wang, P.; Zhang, H.; Tao, Z.; Hou, P.; et al. Continuous detection and genetic diversity of human rotavirus A in sewage in eastern China, 2013–2014. Virol. J. 2016, 13, 153. [Google Scholar] [CrossRef] [Green Version]
- Calgua, B.; Rodriguez-Manzano, J.; Hundesa, A.; Sunen, E.; Calvo, M.; Bofill-Mas, S.; Girones, R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J. Virol. Methods 2013, 187, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Gonzales-Gustavson, E.; Cardenas-Youngs, Y.; Calvo, M.; da Silva, M.F.; Hundesa, A.; Amoros, I.; Moreno, Y.; Moreno-Mesonero, L.; Rosell, R.; Ganges, L.; et al. Characterization of the efficiency and uncertainty of skimmed milk flocculation for the simultaneous concentration and quantification of water-borne viruses, bacteria and protozoa. J. Microbiol. Methods 2017, 134, 46–53. [Google Scholar] [CrossRef]
- Petterson, S.; Grondahl-Rosado, R.; Nilsen, V.; Myrmel, M.; Robertson, L.J. Variability in the recovery of a virus concentration procedure in water: Implications for QMRA. Water Res. 2015, 87, 79–86. [Google Scholar] [CrossRef]
- Levy, K.; Hubbard, A.E.; Eisenberg, J.N. Seasonality of rotavirus disease in the tropics: A systematic review and meta-analysis. Int. J. Epidemiol. 2009, 38, 1487–1496. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.M.; Pitzer, V.E.; Alonso, W.J.; Vera, D.; Lopman, B.; Tate, J.; Viboud, C.; Parashar, U.D. Global seasonality of rotavirus disease. Pediatr. Infect. Dis. J. 2013, 32, e134–e147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villena, C.; El-Senousy, W.M.; Abad, F.X.; Pinto, R.M.; Bosch, A. Group A rotavirus in sewage samples from Barcelona and Cairo: Emergence of unusual genotypes. Appl. Environ. Microbiol. 2003, 69, 3919–3923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthijnssens, J.; Bilcke, J.; Ciarlet, M.; Martella, V.; Banyai, K.; Rahman, M.; Zeller, M.; Beutels, P.; Van Damme, P.; Van Ranst, M. Rotavirus disease and vaccination: Impact on genotype diversity. Future Microbiol. 2009, 4, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, A.; Kuga, K.; Suzuki, T.; Kohmoto, M.; Katsuda, K.; Tsunemitsu, H. Annual changes in predominant genotypes of rotavirus A detected in the feces of pigs in various developmental stages raised on a conventional farm. Vet. Microbiol. 2013, 163, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Papp, H.; Borzak, R.; Farkas, S.; Kisfali, P.; Lengyel, G.; Molnar, P.; Melegh, B.; Matthijnssens, J.; Jakab, F.; Martella, V.; et al. Zoonotic transmission of reassortant porcine G4P[6] rotaviruses in Hungarian pediatric patients identified sporadically over a 15 year period. Infect. Genet. Evol. 2013, 19, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Girones, R.; Ferrus, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; Correa Ade, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water--the pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef]
- Vieira, C.B.; Mendes, A.C.; Guimaraes, F.R.; Fumian, T.M.; Leite, J.P.; Gaspar, A.M.; Miagostovich, M.P. Detection of enteric viruses in recreational waters of an urban lagoon in the city of Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2012, 107, 778–784. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, F.F.; Guimaraes, F.R.; Fumian, T.M.; Victoria, M.; Vieira, C.B.; Luz, S.; Shubo, T.; Leite, J.P.; Miagostovich, M.P. Environmental dissemination of group A rotavirus: P-type, G-type and subgroup characterization. Water Sci. Technol. 2009, 60, 633–642. [Google Scholar] [CrossRef]


| Period | HAdV (CG/L) | RVA (RT-PCR U/L) |
|---|---|---|
| Spring 2015 | 9.51 × 104 | 3.96 × 104 |
| Summer 2015 | 1.71 × 105 | - |
| Fall 2015 | 1.98 × 105 | 1.73 × 106 |
| Winter 2015 | 6.50 × 105 | 1.41 × 107 |
| Spring 2016 | 1.16 × 106 | 3.30 × 108 |
| HAdV | RVA | |||||
|---|---|---|---|---|---|---|
| Summer | Fall | Winter | Summer | Fall | Winter | |
| Fall | 0.97538 | 0.0743 | ||||
| Winter | 0.00077 | 2.0 × 10−8 | 3.6 × 10−6 | 0.0011 | ||
| Spring | 0.00941 | 1.6 × 10−5 | 0.93618 | 1.3 × 10−8 | 2.8 × 10−6 | 0.4376 |
| Season | RVA Genotypes | ||
|---|---|---|---|
| VP4 | VP7 | VP6 | |
| Spring 2015 | P[4] | I2 | |
| Summer 2015 | |||
| Fall 2015 | P[4], P[8], P[9], P[10] | I2 | |
| Winter 2015 | P[4], P[8], P[9] | I1 | |
| Spring 2016 | P[4], P[8] | G2, G3, G9, G12 | I1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Sales, M.; Martínez-Puchol, S.; Gonzales-Gustavson, E.; Hundesa, A.; Gironès, R. High Prevalence of Rotavirus A in Raw Sewage Samples from Northeast Spain. Viruses 2020, 12, 318. https://doi.org/10.3390/v12030318
Silva-Sales M, Martínez-Puchol S, Gonzales-Gustavson E, Hundesa A, Gironès R. High Prevalence of Rotavirus A in Raw Sewage Samples from Northeast Spain. Viruses. 2020; 12(3):318. https://doi.org/10.3390/v12030318
Chicago/Turabian StyleSilva-Sales, Marcelle, Sandra Martínez-Puchol, Eloy Gonzales-Gustavson, Ayalkibet Hundesa, and Rosina Gironès. 2020. "High Prevalence of Rotavirus A in Raw Sewage Samples from Northeast Spain" Viruses 12, no. 3: 318. https://doi.org/10.3390/v12030318
APA StyleSilva-Sales, M., Martínez-Puchol, S., Gonzales-Gustavson, E., Hundesa, A., & Gironès, R. (2020). High Prevalence of Rotavirus A in Raw Sewage Samples from Northeast Spain. Viruses, 12(3), 318. https://doi.org/10.3390/v12030318