Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. A Study of New Symptomatic Tomato Plants in Israel and ToBRFV and/or PepMV Inoculation Experiments
2.2. Infectious Potential
2.3. PepMV Virion Purification and Specific Antiserum Preparation
2.4. Indirect Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. Viral RNA Extraction, Reverse Transcription (RT) and PCR Amplification
2.6. High-Throughput Sequencing (HTS) with Illumina HiSeq Platform and Bioinformatic Analysis
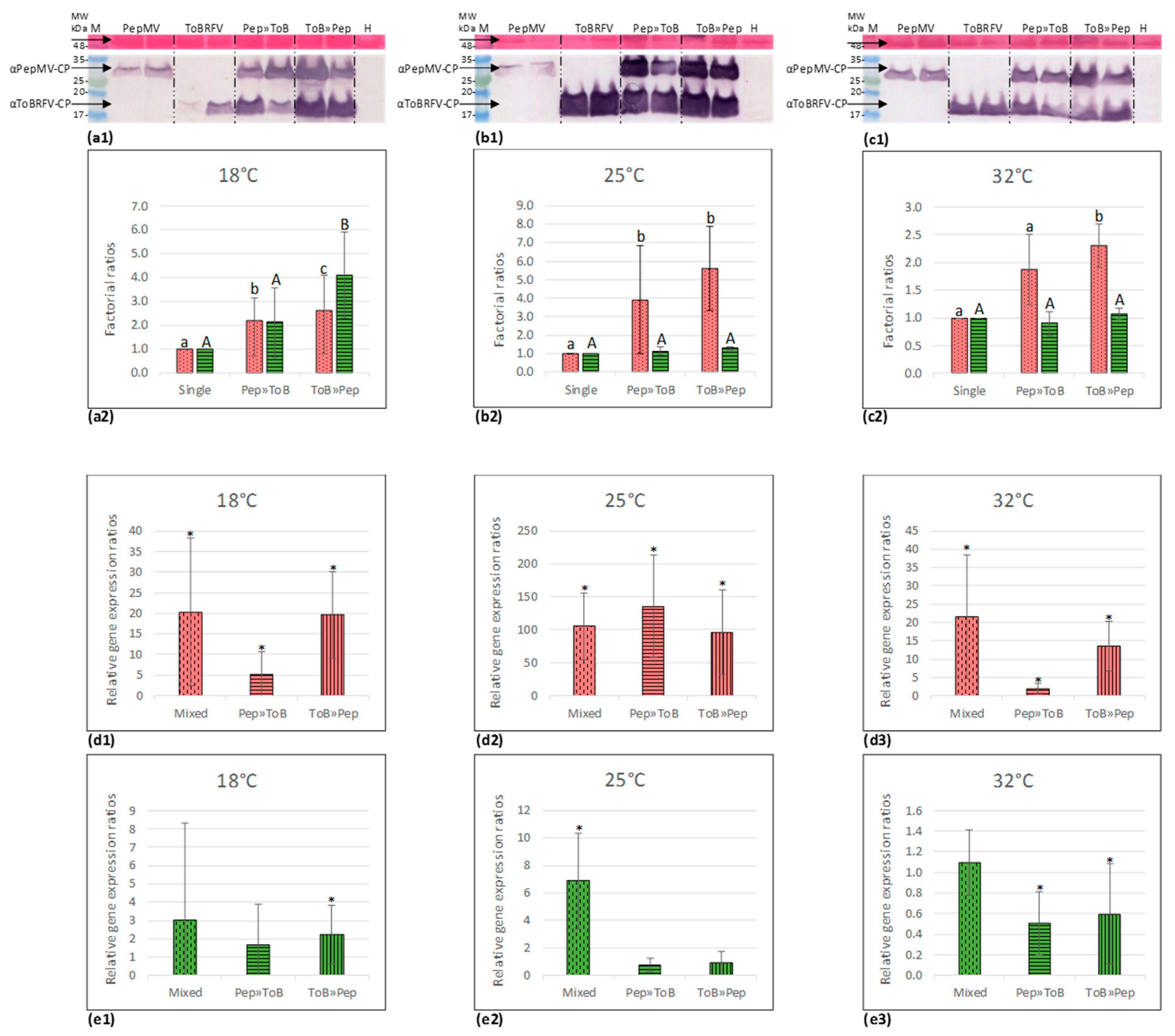
2.7. Western Blot Analyses
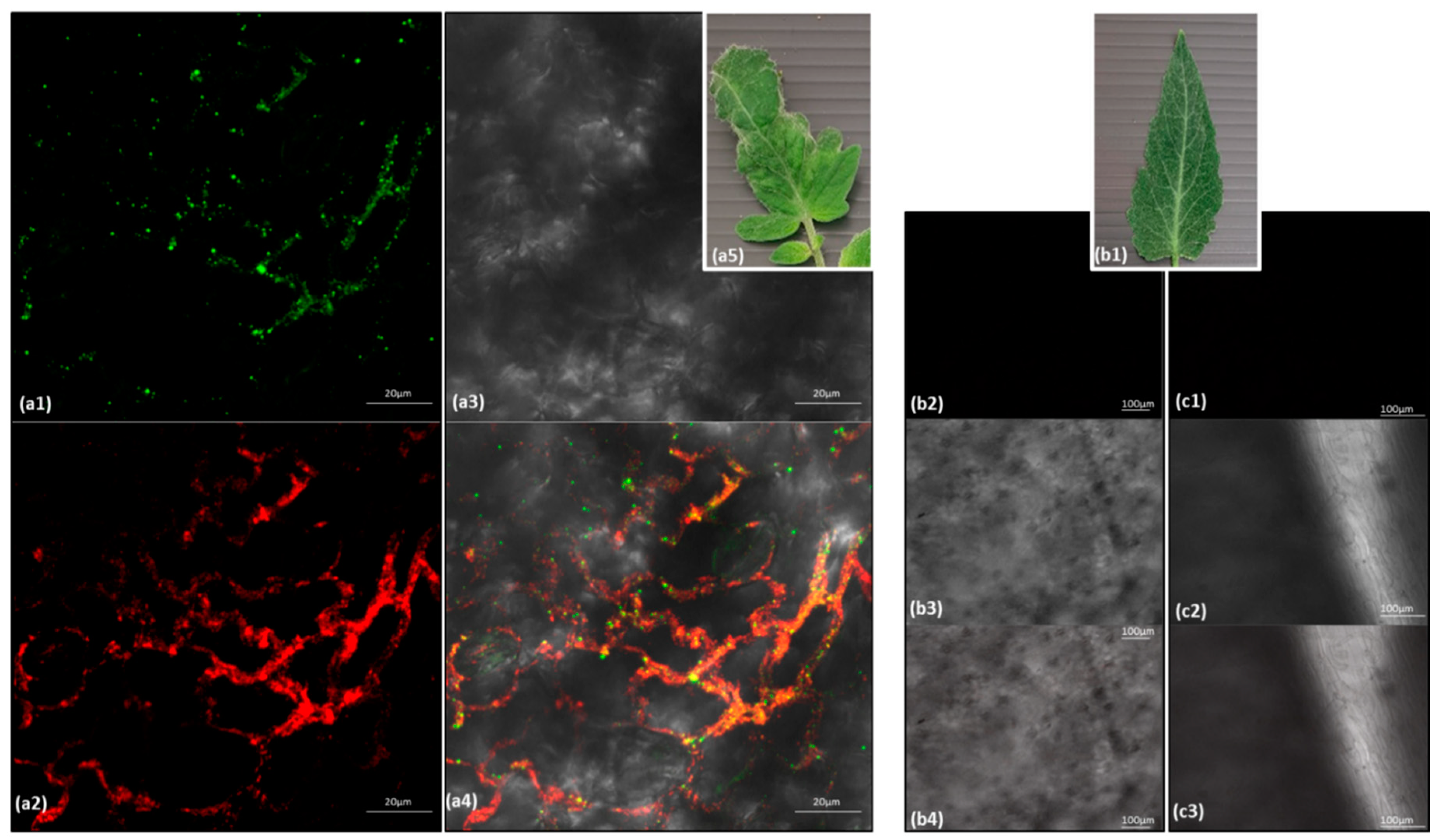
2.8. In Situ Immunofluorescence
2.9. Quantitative RT-PCR (RT-qPCR)
2.10. Synergy Factor (SF) Calculations
3. Results
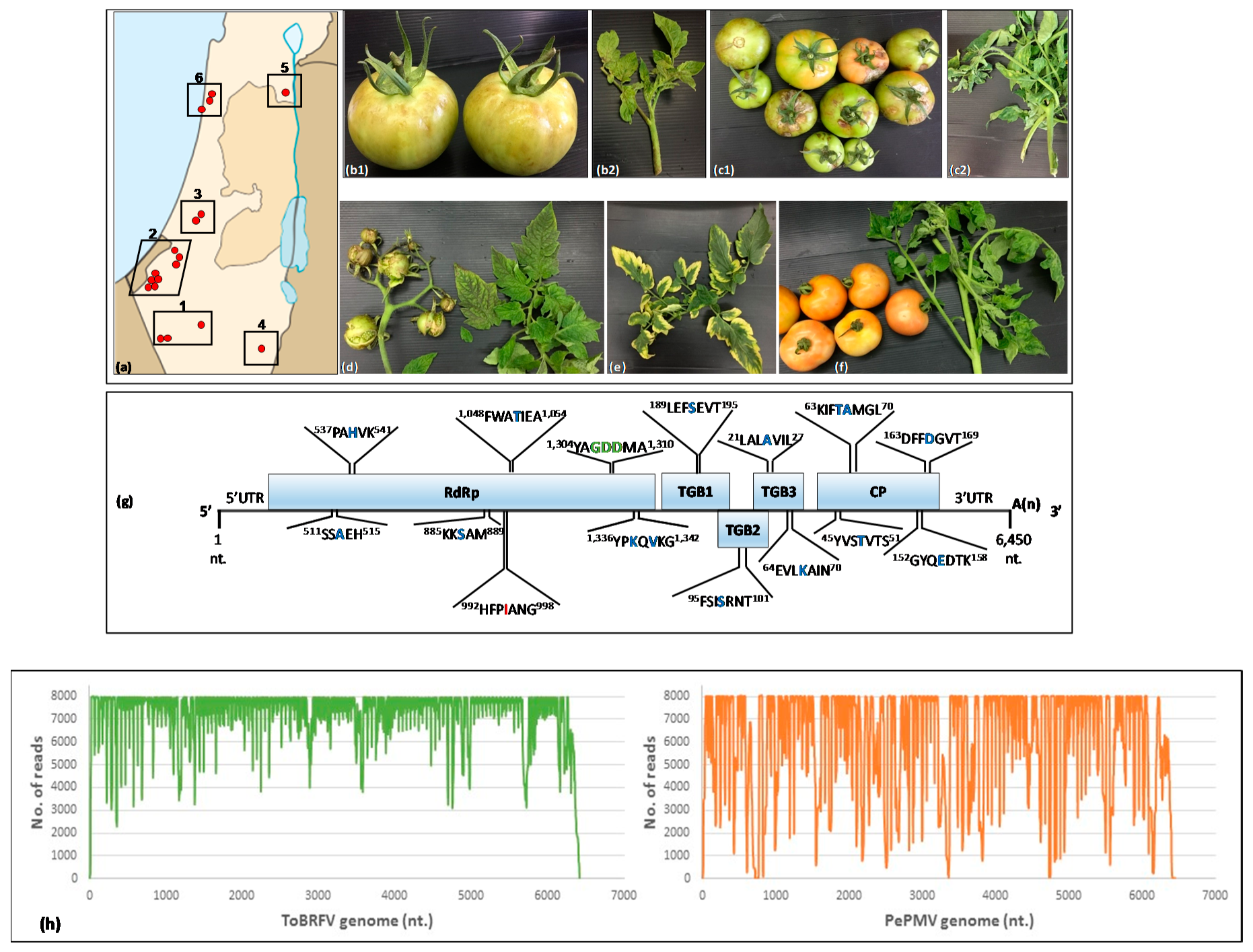
3.1. ToBRFV and PepMV Co-Infections in Distinctly Severe Symptomatic Tomato Plants
3.2. Genome Characterization of PepMV Isolates in Symptomatic Tomato Plants Co-Infected by ToBRFV and PepMV
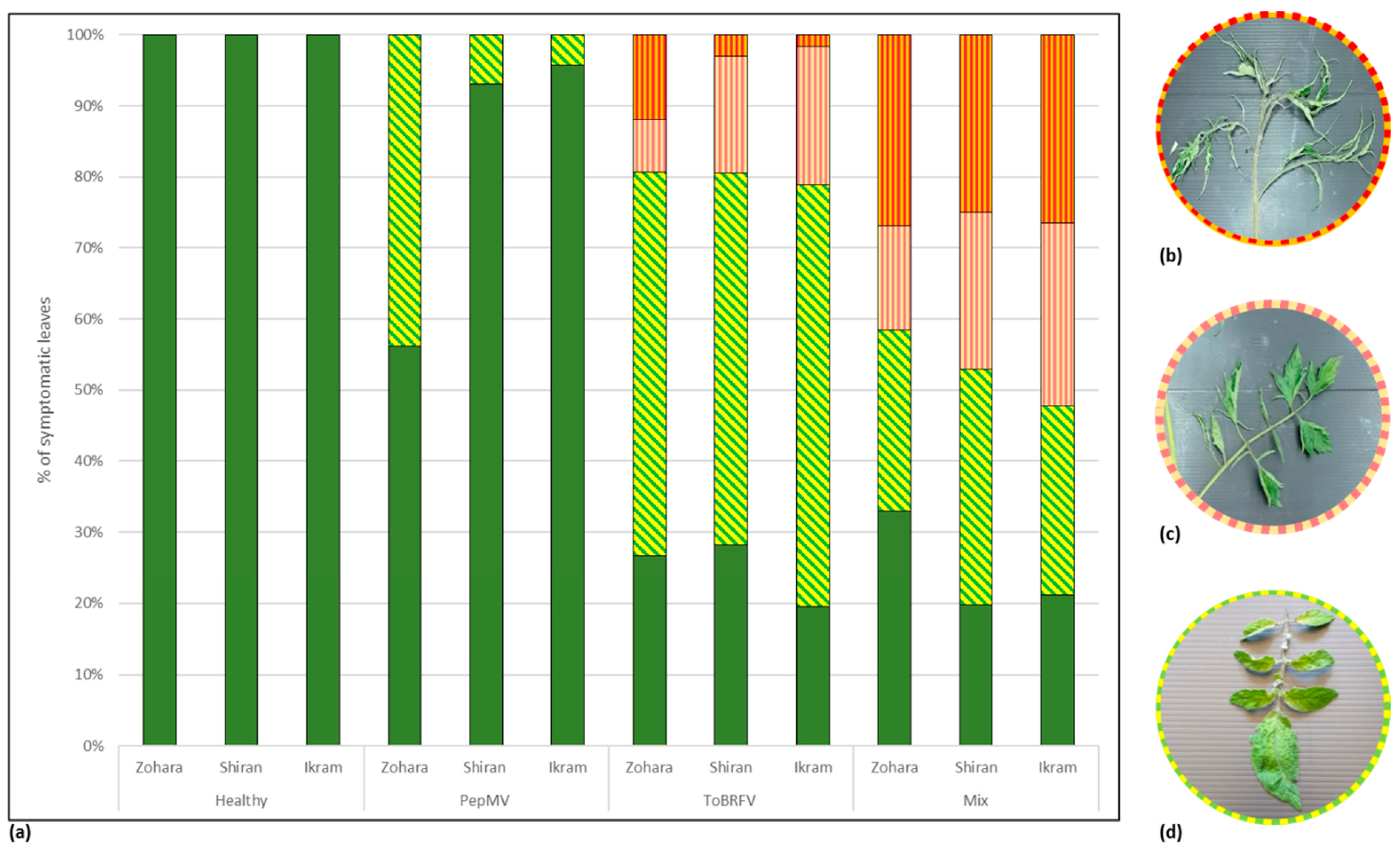
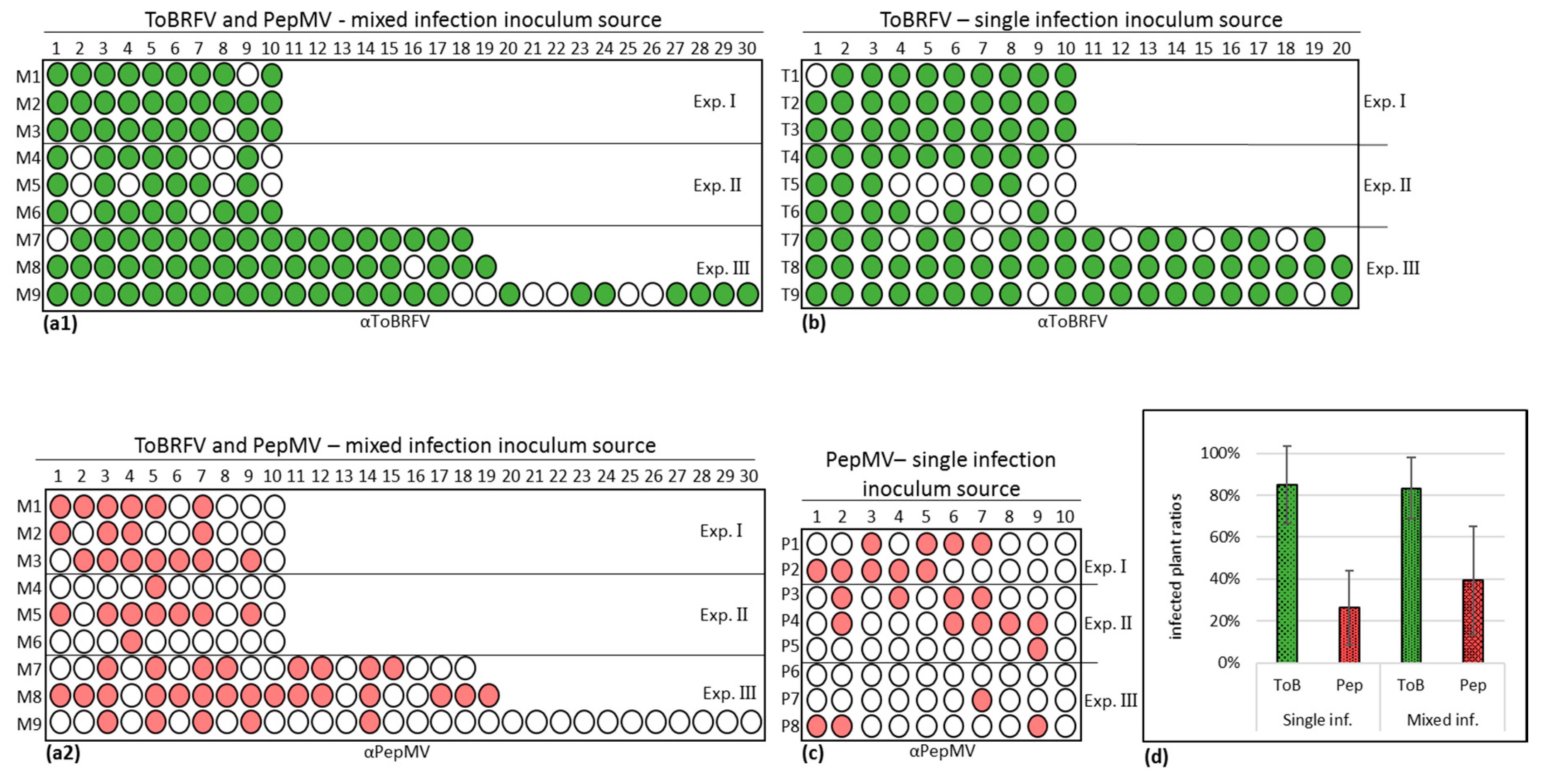
3.3. Biological Assays and Serological Tests Showing PepMV Mild Effects in Singly Infected Tomato Plants Turn Aggressive in Mixed Infections with ToBRFV
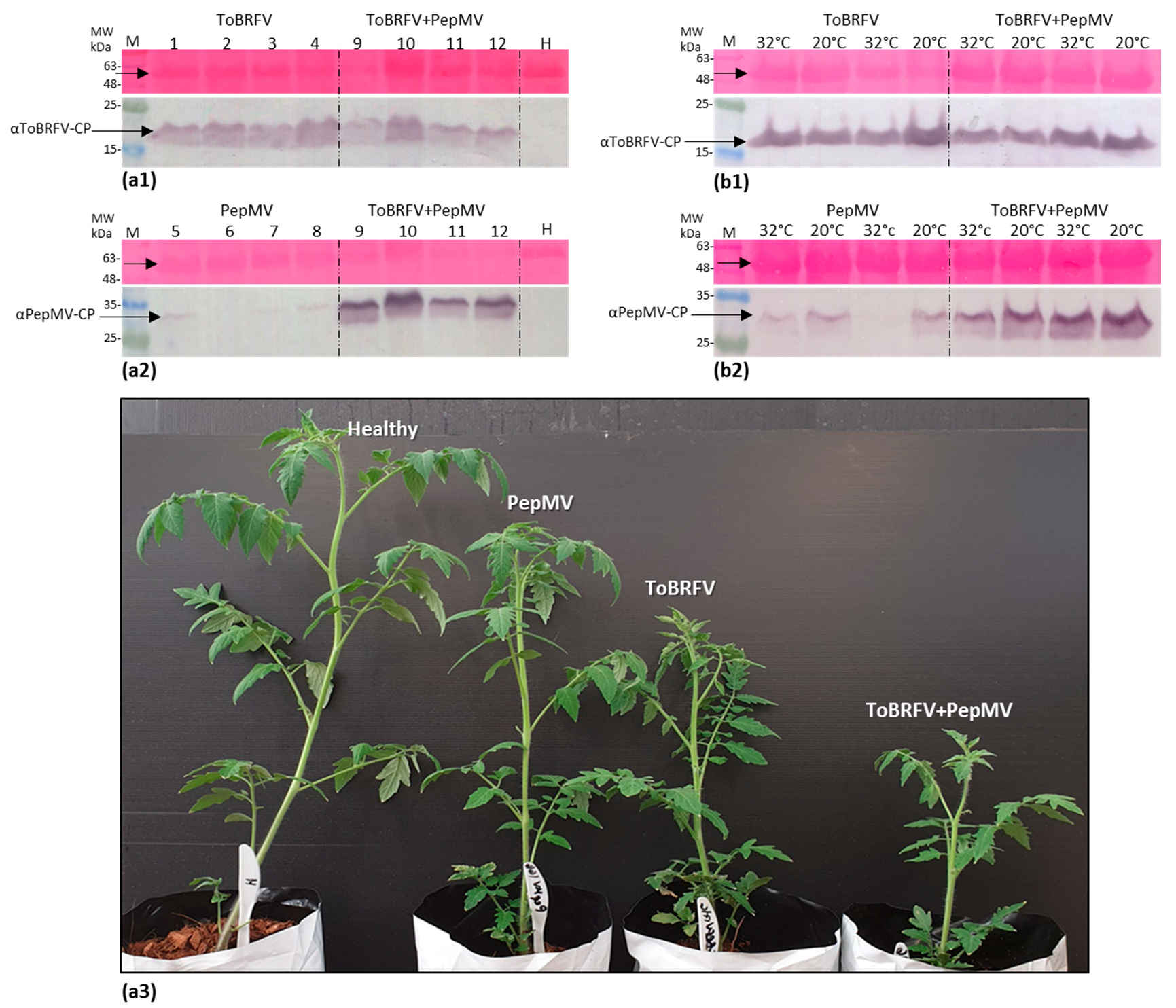
3.4. PepMV Inoculations of Tomato Plants, Preceded by ToBRFV, Resulted in a High PepMV Titer Compared to PepMV Single Inoculations
3.5. Synergy Factor (SF) for ToBRFV and PepMV Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A New Israeli Tobamovirus Isolate Infects Tomato Plants Harboring Tm-22 Resistance Genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [Green Version]
- Fidan, H.; Sarikaya, P.; Calis, O. First report of Tomato brown rugose fruit virus on tomato in Turkey. New Dis. Rep. 2019, 39, 18. [Google Scholar] [CrossRef] [Green Version]
- Menzel, W.; Knierim, D.; Winter, S.; Hamacher, J.; Heupel, M. First report of tomato brown rugose fruit virus infecting tomato in Germany. New Dis. Rep. 2019, 39, 2044. [Google Scholar] [CrossRef] [Green Version]
- Panno, S.; Caruso, A.; Davino, S. First Report of Tomato Brown Rugose Fruit Virus on Tomato Crops in Italy. Plant Dis. 2019, 103, 1443. [Google Scholar] [CrossRef]
- Skelton, A.; Buxton-Kirk, A.; Ward, R.; Harju, V.; Frew, L.; Fowkes, A.; Long, M.; Negus, A.; Forde, S.; Adams, I. First report of Tomato brown rugose fruit virus in tomato in the United Kingdom. New Dis. Rep. 2019, 40, 12. [Google Scholar] [CrossRef] [Green Version]
- Cambrón-Crisantos, J.M.; Rodríguez-Mendoza, J.; Valencia-Luna, J.B.; Rangel, S.A.; de Jesús García-Ávila, C.; López-Buenfil, J.A. First report of Tomato brown rugose fruit virus (ToBRFV) in Michoacan, Mexico. Mex. J. Phytopathol. 2018, 37, 185–192. [Google Scholar]
- Ling, K.-S.; Tian, T.; Gurung, S.; Salati, R.; Gilliard, A. First report of tomato brown rugose fruit virus infecting greenhouse tomato in the US. Plant Dis. 2019, 103, 1439. [Google Scholar] [CrossRef]
- Yan, Z.; Ma, H.; Han, S.; Geng, C.; Tian, Y.; Li, X. First report of Tomato brown rugose fruit virus infecting tomato in China. Plant Dis. 2019, 103, 2973. [Google Scholar] [CrossRef]
- Beris, D.; Malandraki, I.; Kektsidou, O.; Theologidis, I.; Vassilakos, N.; Varveri, C. First report of Tomato brown rugose fruit virus infecting tomato in Greece. Plant Dis. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wright, D.; Mumford, R. Pepino mosaic Potexvirus (PepMV): First records in tomato in the United Kingdom. In Plant Disease Notice; Central Science Laboratory: York, UK, 1999; Volume 89, p. 400. [Google Scholar]
- Van der Vlugt, R.A.A.; Stijger, C.C.M.M.; Verhoeven, J.T.J.; Lesemann, D.E. First report of Pepino mosaic virus on tomato. Plant Dis. 2000, 84, 103. [Google Scholar] [CrossRef] [PubMed]
- Lesemann, D.-E.; Dalchow, J.; Winter, S.; Pfeilstetter, E. Occurrence of Pepino mosaic virus in European tomato crops: Identification, etiology and epidemiology. Mitt. -Biol. Bundesanst. Fur Land Und Forstwirtsch. 2000, 323. [Google Scholar]
- French, C.; Bouthillier, M.; Bernardy, M.; Ferguson, G.; Sabourin, M.; Johnson, R.; Masters, C.; Godkin, S.; Mumford, R. First report of Pepino mosaic virus in Canada and the United States. Plant Dis. 2001, 85, 1121. [Google Scholar] [CrossRef] [PubMed]
- Jordá, C.; Perez, A.L.; Martínez-Culebras, P.; Abad, P.; Lacasa, A.; Guerrero, M. First report of Pepino mosaic virus on tomato in Spain. Plant Dis. 2001, 85, 1292. [Google Scholar] [CrossRef] [PubMed]
- Mumford, R.; Metcalfe, E. The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch. Virol. 2001, 146, 2455–2460. [Google Scholar] [CrossRef] [PubMed]
- Roggero, P.; Masenga, V.; Lenzi, R.; Coghe, F.; Ena, S.; Winter, S. First report of Pepino mosaic virus in tomato in Italy. Plant Pathol. 2001, 50, 798. [Google Scholar] [CrossRef]
- Klap, C.; Luria, N.; Smith, E.; Bakelman, E.; Belausov, E.; Laskar, O.; Lachman, O.; Gal-On, A.; Dombrovsky, A. The Potential Risk of Plant-Virus Disease Initiation by Infected Tomatoes. Plants 2020, 9, 623. [Google Scholar] [CrossRef]
- Jones, R.A.C.; Koening, R.; Lesemann, D.E. Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum). Ann. Appl. Biol. 1980, 94, 61–68. [Google Scholar] [CrossRef]
- Ling, K.-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007, 34, 1–8. [Google Scholar] [CrossRef]
- Maroon-Lango, C.; Guaragna, M.; Jordan, R.; Hammond, J.; Bandla, M.; Marquardt, S. Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European-like) US isolate. Arch. Virol. 2005, 150, 1187–1201. [Google Scholar] [CrossRef]
- Gómez, P.; Sempere, R.; Elena, S.F.; Aranda, M.A. Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J. Virol. 2009, 83, 12378–12387. [Google Scholar] [CrossRef] [Green Version]
- Hasiów-Jaroszewska, B.; Pospieszny, H.; Borodynko, N. New necrotic isolates of Pepino mosaic virus representing the CH2 genotype. J. Phytopathol. 2009, 157, 494–496. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Thomma, B.P. Pepino mosaic virus: A successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol. Plant Pathol. 2010, 11, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, I.; Paeleman, A.; Vandewoestijne, E.; Van Bergen, L.; Bragard, C.; Lievens, B.; Vanachter, A.; Thomma, B. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol. 2009, 58, 450–460. [Google Scholar] [CrossRef]
- Spence, N.; Basham, J.; Mumford, R.; Hayman, G.; Edmondson, R.; Jones, D. Effect of Pepino mosaic virus on the yield and quality of glasshouse-grown tomatoes in the UK. Plant Pathol. 2006, 55, 595–606. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Gutiérrez-Aguirre, I.; Paeleman, A.; Goen, K.; Wittemans, L.; Lievens, B.; Vanachter, A.C.; Ravnikar, M.; Thomma, B. Cross-protection or enhanced symptom display in greenhouse tomato co-infected with different Pepino mosaic virus isolates. Plant Pathol. 2010, 59, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Sempere, R.N.; Gómez-Aix, C.; Ruíz-Ramón, F.; Gómez, P.; Hasiów-Jaroszewska, B.; Sánchez-Pina, M.A.; Aranda, M.A. Pepino mosaic virus RNA-dependent RNA polymerase pol domain is a hypersensitive response-like elicitor shared by necrotic and mild isolates. Phytopathology 2016, 106, 395–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasiów-Jaroszewska, B.; Borodynko, N. Characterization of the necrosis determinant of the European genotype of pepino mosaic virus by site-specific mutagenesis of an infectious cDNA clone. Arch. Virol. 2012, 157, 337–341. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Jackowiak, P.; Figlerowicz, M.; Pospieszny, H. Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res. 2011, 159, 57–61. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Paeleman, A.; Ortega-Parra, N.; Borodynko, N.; Minicka, J.; Czerwoniec, A.; Thomma, B.P.; Hanssen, I.M. Ratio of mutated versus wild-type coat protein sequences in P epino mosaic virus determines the nature and severity of yellowing symptoms on tomato plants. Mol. Plant Pathol. 2013, 14, 923–933. [Google Scholar] [CrossRef]
- Duff-Farrier, C.R.; Bailey, A.M.; Boonham, N.; Foster, G.D. A pathogenicity determinant maps to the N-terminal coat protein region of the P epino mosaic virus genome. Mol. Plant Pathol. 2015, 16, 308–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chewachong, G.M.; Miller, S.A.; Blakeslee, J.J.; Francis, D.M.; Morris, T.J.; Qu, F. Generation of an attenuated, cross-protective Pepino mosaic virus variant through alignment-guided mutagenesis of the viral capsid protein. Phytopathology 2015, 105, 126–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blystad, D.-R.; van der Vlugt, R.; Alfaro-Fernández, A.; del Carmen Córdoba, M.; Bese, G.; Hristova, D.; Pospieszny, H.; Mehle, N.; Ravnikar, M.; Tomassoli, L. Host range and symptomatology of Pepino mosaic virus strains occurring in Europe. Eur. J. Plant Pathol. 2015, 143, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Gao, S.; Padmanabhan, C.; Li, R.; Galvez, M.; Gutierrez, D.; Fuentes, S.; Ling, K.-S.; Kreuze, J.; Fei, Z. VirusDetect: An automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 2017, 500, 130–138. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Lacerda, A.L.; Fonseca, L.N.; Blawid, R.; Boiteux, L.S.; Ribeiro, S.G.; Brasileiro, A.C. Reference gene selection for qPCR analysis in tomato-bipartite begomovirus interaction and validation in additional tomato-virus pathosystems. PLoS ONE 2015, 10, e0136820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical analysis of real-time PCR data. Bmc Bioinform. 2006, 7, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Abbott, W.S. A method of computing the effectiveness of an insecticide. J. Econ. Entomol 1925, 18, 265–267. [Google Scholar] [CrossRef]
- Murphy, J.F.; Bowen, K.L. Synergistic disease in pepper caused by the mixed infection of Cucumber mosaic virus and Pepper mottle virus. Phytopathology 2006, 96, 240–247. [Google Scholar] [CrossRef] [Green Version]
- Levy, Y.; Benderly, M.; Cohen, Y.; Gisi, U.; Bassand, D. The joint action of fungicides in mixtures: Comparison of two methods for synergy calculation. Eppo Bull. 1986, 16, 651–657. [Google Scholar] [CrossRef]
- Hu, W.-W.; Wong, S.-M.; Loh, C.-S.; Goh, C.-J. Synergism in replication of cymbidium mosaic potexvirus (CymMV) and odontoglossum ringspot tobamovirus (ORSV) RNA in orchid protoplasts. Arch. Virol. 1998, 143, 1265–1275. [Google Scholar] [CrossRef]
- SYu, M.; Fedorkin, O.; Schiemann, J.; Baulcombe, D.; Atabekov, J. Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J. Gen. Virol. 1997, 78, 2077–2083. [Google Scholar]
- Ajjikuttira, P.; Loh, C.-S.; Wong, S.-M. Reciprocal function of movement proteins and complementation of long-distance movement of Cymbidium mosaic virus RNA by Odontoglossum ringspot virus coat protein. J. Gen. Virol. 2005, 86, 1543–1553. [Google Scholar] [CrossRef]
- MacNeill, B.H.; Ismen, H. Studies on the virus-streak syndrome in tomatoes. Can. J. Bot. 1960, 38, 9–20. [Google Scholar] [CrossRef]
- Murakishi, H.; Honma, S. Resistance to tobacco mosaic virus in Lycopersicon hybrids evaluated by potato virus X synergy. Euphytica 1963, 12, 27–31. [Google Scholar]
- Balogun, O.S.; Xu, L.; Teraoka, T.; Hosokawa, D. Effects of single and double infections with Potato virus X and Tobacco mosaic virus on disease development, plant growth, and virus accumulation in tomato. Fitopatol. Bras. 2002, 27, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Balogun, O.S. Seedling age at inoculation and infection sequence affect disease and growth response in tomato mixed infected with potato virus X and tomato mosaic virus. Int. J. Agric. Biol. 2008, 10, 145–150. [Google Scholar]
- Syller, J. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.M.; Ross, A.F. Enhancement by potato virus Y of potato virus X synthesis in doubly infected tobacco depends on the timing of invasion by the viruses. Virology 1974, 58, 263–271. [Google Scholar] [CrossRef]
- Chávez-Calvillo, G.; Contreras-Paredes, C.A.; Mora-Macias, J.; Noa-Carrazana, J.C.; Serrano-Rubio, A.A.; Dinkova, T.D.; Carrillo-Tripp, M.; Silva-Rosales, L. Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology 2016, 489, 179–191. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.S.; Liu, J.; Cheng, N.-H.; Folimonov, A.; Hou, Y.-M.; Bao, Y.; Katagi, C.; Carter, S.A.; Nelson, R.S. The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol. Plant-Microbe Interact. 2004, 17, 583–592. [Google Scholar] [CrossRef] [Green Version]
- Vogler, H.; Akbergenov, R.; Shivaprasad, P.V.; Dang, V.; Fasler, M.; Kwon, M.-O.; Zhanybekova, S.; Hohn, T.; Heinlein, M. Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J. Virol. 2007, 81, 10379–10388. [Google Scholar] [CrossRef] [Green Version]
- Kubota, K.; Tsuda, S.; Tamai, A.; Meshi, T. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 2003, 77, 11016–11026. [Google Scholar] [CrossRef] [Green Version]
- Palauqui, J.C.; Elmayan, T.; Pollien, J.M.; Vaucheret, H. Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. Embo J. 1997, 16, 4738–4745. [Google Scholar] [CrossRef]
- Voinnet, O.; Baulcombe, D.C. Systemic signalling in gene silencing. Nature 1997, 389, 553. [Google Scholar] [CrossRef] [PubMed]
- Moissiard, G.; Voinnet, O. Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 2004, 5, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Pruss, G.; Ge, X.; Shi, X.M.; Carrington, J.C.; Vance, V.B. Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 1997, 9, 859–868. [Google Scholar] [CrossRef]
- Schwach, F.; Vaistij, F.E.; Jones, L.; Baulcombe, D.C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005, 138, 1842–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayne, E.H.; Rakitina, D.V.; Morozov, S.Y.; Baulcombe, D.C. Cell-to-cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant J. 2005, 44, 471–482. [Google Scholar] [CrossRef]
- Pacheco, R.; García-Marcos, A.; Barajas, D.; Martiáñez, J.; Tenllado, F. PVX–potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res. 2012, 165, 231–235. [Google Scholar] [CrossRef]
- Scholthof, H.B. The Tombusvirus-encoded P19: From irrelevance to elegance. Nat. Rev. Microbiol. 2006, 4, 405–411. [Google Scholar] [CrossRef]
- Andrade, O.; Latorre, B.; Escaffi, O. Tomato mosaic virus associated with shoestring symptom in Chilean tomatoes. Plant Dis. 1981, 65, 761–762. [Google Scholar] [CrossRef]
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier Academic Press: San Diego, CA, USA, 2005. [Google Scholar]
- Mascia, T.; Gallitelli, D. Synergies and antagonisms in virus interactions. Plant Sci. 2016, 252, 176–192. [Google Scholar] [CrossRef]
| Set No. | Orientation | Name (nt. Position) | Amplicon Length (bp) | Sequence (5′-3′) |
|---|---|---|---|---|
| 1 | F | ToBRFV (5557) | 615 | TTTAGTAGTAAAAGTGAGAAT |
| RC | ToBRFV (6167) | TTGTAAACCGGATGCACTTTCAAATG | ||
| 2 | F | PepMV (5658) | 650 | CCATCAGATGCACCACCAAC |
| RC | PepMV (6307) | TTAGCTCCTCCCATGTGTCC | ||
| 3 | F | PepMV (1) | 1184 | GAAAACAAAACATAACACATAATA |
| RC | PepMV (1184) | AAAAACTTGTCGCACCCATG | ||
| 4 | F | PepMV (925) | 1070 | AACATCTTCCATCCCCAACA |
| RC | PepMV (1994) | CGTCTTCTCTGCCATGTGAA | ||
| 5 | F | PepMV (1711) | 1164 | TGGGCTAGTCTTGCATCTGA |
| RC | PepMV (2874) | GTCTCGTGCAGTTGATTCCA | ||
| 6 | F | PepMV (2698) | 1205 | CAAAAGTGCAAAGTGCCAAT |
| RC | PepMV (3902) | CTGGTGAAGGTCCCACATTT | ||
| 7 | F | PepMV (3619) | 1097 | TTAAGGAAAATGCGGCAAAG |
| RC | PepMV (4715) | ATTTTTGGTGACCCCTGTCA | ||
| 8 | F | PepMV (4522) | 1130 | AACTGGGAAAACCACATTGC |
| RC | PepMV (5651) | CTAACCCATCAGATGCACCA | ||
| 9 | F | PepMV (6075) | 340 | GCAAAATTGGGCTATCAGGA |
| RC | PepMV (6414) | ATTTAGTAGATTTAGATACTAAGG |
| Location (Number of Samples) | ELISA | a RT-PCR | ||
|---|---|---|---|---|
| ToBRFV | PepMV | ToBRFV | PepMV | |
| 1. Ramat Negev (11) | 11/11 | 10/11 | 11 | 10 |
| 2. Bsor (69) | 69/69 | 50/69 | 69 | 50 |
| 3. Lakhish (5) | 5/5 | 5/5 | 5 | 3 |
| 4. Arava (5) | 5/5 | 5/5 | 5 | 2 |
| 5. Beit She’an Valley (8) | 5/8 | 5/8 | 5 | 1 |
| 6. Hefer Valley (6) | 6/6 | 6/6 | 6 | 6 |
| Temp. | PepMV | ToBRFV | Mixed | PepMV »ToBRFV | ToBRFV »PepMV |
|---|---|---|---|---|---|
| 18 °C | LY (20–30%) LM (20–30%) | LY (20–30%) LM (80–90%) LD (20–30%) | LY (20–30%) LM (80–90%) LD (80–90%) | LY (60–70%) LM (60–70%) LD (60–70%) | LY (80–90%) LM (80–90%) LD (80–90%) |
| 25 °C | NS | LM (60–70%) LD (20–30%) | LY (60–70%) LM (60–70%) LD (60–70%) | LY (20–30%) LM (80–90%) LD (60–70%) | LY (60–70%) LM (60–70%) LD (20–30%) |
| 32 °C | NS | LM (20–30%) LD (60–70%) | LM (60–70%) LD (20–30%) | LY (60–70%) LM (60–70%) LD (60–70%) | LY (20–30%) LM (20–30%) LD (60–70%) |
| Treatment | Temp. | Mean Heights (cm) | s.d. (n = 4) | % Height Reductions | SF | |
| Healthy | 18 | 77 | 7.39 | 0% | - | |
| Mixed | 18 | 46.75 | 2.5 | 39% | 1.10 | |
| PepMV»ToBRFV | 18 | 43.25 | 2.22 | 44% | 1.23 | |
| PepMV | 18 | 52.5 | 2.38 | 32% | - | |
| ToBRFV | 18 | 74 | 1.63 | 4% | - | |
| Treatment | Temp. | Mean Number of Leaves | s.d. | % Leaf Reductions | SF | |
| Healthy | 18 | 10.75 | 0.5 | 0% | - | |
| Mixed | 18 | 10 | 0.82 | 7% | 2.99 | |
| PepMV»ToBRFV | 18 | 9 | 0.82 | 16% | 6.97 | |
| PepMV | 18 | 9.5 | 1.29 | 12% | - | |
| ToBRFV | 18 | 11.75 | 1.5 | −9% | - | |
| Treatment | Temp. | Mean Intensity Ratios, PepMV | s.d. | Mean Intensity Ratios ToBRFV | s.d. | SF |
| PepMV | 18 °C | 76% | 35% | - | - | - |
| ToBRFV | 18 °C | - | - | 64% | 49% | - |
| PepMV»ToBRFV | 18 °C | 125% | 33% | 120% | 42% | 1.75 |
| ToBRFV»PepMV | 18 °C | 190% | 106% | 250% | 95% | 3.14 |
| PepMV | 25–45 °C | 20% | 3% | - | - | - |
| ToBRFV | 25–45 °C | - | - | 117% | 37% | - |
| Mixed | 25–45 °C | 123% | 31% | 78% | 18% | 1.47 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klap, C.; Luria, N.; Smith, E.; Hadad, L.; Bakelman, E.; Sela, N.; Belausov, E.; Lachman, O.; Leibman, D.; Dombrovsky, A. Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants. Viruses 2020, 12, 879. https://doi.org/10.3390/v12080879
Klap C, Luria N, Smith E, Hadad L, Bakelman E, Sela N, Belausov E, Lachman O, Leibman D, Dombrovsky A. Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants. Viruses. 2020; 12(8):879. https://doi.org/10.3390/v12080879
Chicago/Turabian StyleKlap, Chen, Neta Luria, Elisheva Smith, Lior Hadad, Elena Bakelman, Noa Sela, Eduard Belausov, Oded Lachman, Diana Leibman, and Aviv Dombrovsky. 2020. "Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants" Viruses 12, no. 8: 879. https://doi.org/10.3390/v12080879
APA StyleKlap, C., Luria, N., Smith, E., Hadad, L., Bakelman, E., Sela, N., Belausov, E., Lachman, O., Leibman, D., & Dombrovsky, A. (2020). Tomato Brown Rugose Fruit Virus Contributes to Enhanced Pepino Mosaic Virus Titers in Tomato Plants. Viruses, 12(8), 879. https://doi.org/10.3390/v12080879






